Molecular diagnostic approach to non-Hodgkin's lymphoma
- PMID: 11232108
- PMCID: PMC1906917
- DOI: 10.1016/S1525-1578(10)60636-8
Molecular diagnostic approach to non-Hodgkin's lymphoma
Figures

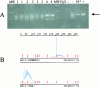


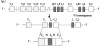
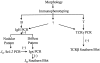
References
-
- Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 - PubMed
-
- Harris NL, Jaffe ES, Diebold J, Flandrin G, Müller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD: The World Health Organization Classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the clinical advisory committee meeting, Airlie House, Virginia, November 1997. J Clin Oncol 1997, 17:3835-3849 - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM: Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403:503-511 - PubMed
-
- Berns A: Gene expression in diagnosis. Nature 2000, 403:491-492 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

