Integration of cytogenetic landmarks into the draft sequence of the human genome
- PMID: 11237021
- PMCID: PMC7845515
- DOI: 10.1038/35057192
Integration of cytogenetic landmarks into the draft sequence of the human genome
Abstract
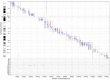
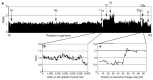
We have placed 7,600 cytogenetically defined landmarks on the draft sequence of the human genome to help with the characterization of genes altered by gross chromosomal aberrations that cause human disease. The landmarks are large-insert clones mapped to chromosome bands by fluorescence in situ hybridization. Each clone contains a sequence tag that is positioned on the genomic sequence. This genome-wide set of sequence-anchored clones allows structural and functional analyses of the genome. This resource represents the first comprehensive integration of cytogenetic, radiation hybrid, linkage and sequence maps of the human genome; provides an independent validation of the sequence map and framework for contig order and orientation; surveys the genome for large-scale duplications, which are likely to require special attention during sequence assembly; and allows a stringent assessment of sequence differences between the dark and light bands of chromosomes. It also provides insight into large-scale chromatin structure and the evolution of chromosomes and gene families and will accelerate our understanding of the molecular bases of human disease and cancer.
Figures

References
-
- Trask BJ. Genome Analysis: A Laboratory Manual. 1999. pp. 303–413.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

