Regulation of CDC42 GTPase by proline-rich tyrosine kinase 2 interacting with PSGAP, a novel pleckstrin homology and Src homology 3 domain containing rhoGAP protein
- PMID: 11238453
- PMCID: PMC2198805
- DOI: 10.1083/jcb.152.5.971
Regulation of CDC42 GTPase by proline-rich tyrosine kinase 2 interacting with PSGAP, a novel pleckstrin homology and Src homology 3 domain containing rhoGAP protein
Abstract
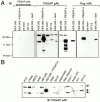
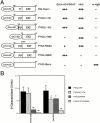
Proline-rich tyrosine kinase 2 (PYK2), a tyrosine kinase structurally related to focal adhesion kinase (FAK), is implicated in regulating cytoskeletal organization. However, mechanisms by which PYK2 participates in and regulates cytoskeletal organization remain largely unknown. Here we report identification of PSGAP, a novel protein that interacts with PYK2 and FAK and contains multiple domains including a pleckstrin homology domain, a rhoGTPase-activating protein domain, and a Src homology 3 domain. PYK2 interacts with PSGAP Src homology 3 domain via the carboxyl-terminal proline-rich sequence. PSGAP is able to increase GTPase activity of CDC42 and RhoA in vitro and in vivo. Remarkably, PYK2, but not FAK, can activate CDC42 via inhibition of PSGAP-mediated GTP hydrolysis of CDC42. Moreover, PSGAP is localized at cell periphery in fibroblasts in a pleckstrin homology domain-dependent manner. Over expression of PSGAP in fibroblasts results in reorganization of cytoskeletal structures and changes of cellular morphology, which requires rhoGTPase-activating activity. Taken together, our results suggest that PSGAP is a signaling protein essential for PYK2 regulation of cytoskeletal organization via Rho family GTPases.
Figures









References
-
- Astier A., Avraham H., Manie S.N., Groopmen J., Canty T., Avraham S., Freedman A.S. The related adhesion focal tyrosine kinase is tyrosine-phosphorylated after β1-integrin stimulation in B cells and binds to p130Cas . J. Biol. Chem. 1997;272:228–232. - PubMed
-
- Avraham S., London R., Fu Y., Ota S., Hiregowdara D., Li J., Jiang S., Pasztor L.M., White R.A., Groopman J.E., Avraham H. Identification and characterization of a novel related adhesion focal tyrosine kinase (RAFTK) from megakaryocytes and brain. J. Biol. Chem. 1995;270:27742–27751. - PubMed
-
- Bar-Sagi D., Rotin D., Batzer A., Mandiyan V., Schlessinger J. SH3 domains direct cellular localization of signaling molecules. Cell. 1993;74:83–91. - PubMed
-
- Barrett T., Xiao B., Dodson E.J., Dodson G., Ludbrook S.B., Nurmahomed K., Gamblin S.J., Musacchio A., Smerdon S.J., Eccleston J.F. The structure of the GTPase-activating domain from p50rhoGAP. Nature. 1997;385:458–461. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

