"Hit-and-run" transformation by adenovirus oncogenes
- PMID: 11238835
- PMCID: PMC114102
- DOI: 10.1128/JVI.75.7.3089-3094.2001
"Hit-and-run" transformation by adenovirus oncogenes
Abstract
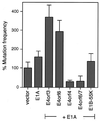
According to classical concepts of viral oncogenesis, the persistence of virus-specific oncogenes is required to maintain the transformed cellular phenotype. In contrast, the "hit-and-run" hypothesis claims that viruses can mediate cellular transformation through an initial "hit," while maintenance of the transformed state is compatible with the loss ("run") of viral molecules. It is well established that the adenovirus E1A and E1B gene products can cooperatively transform primary human and rodent cells to a tumorigenic phenotype and that these cells permanently express the viral oncogenes. Additionally, recent studies have shown that the adenovirus E4 region encodes two novel oncoproteins, the products of E4orf6 and E4orf3, which cooperate with the viral E1A proteins to transform primary rat cells in an E1B-like fashion. Unexpectedly, however, cells transformed by E1A and either E4orf6 or E4orf3 fail to express the viral E4 gene products, and only a subset contain E1A proteins. In fact, the majority of these cells lack E4- and E1A-specific DNA sequences, indicating that transformation occurred through a hit-and-run mechanism. We provide evidence that the unusual transforming activities of the adenoviral oncoproteins may be due to their mutagenic potential. Our results strongly support the possibility that even tumors that lack any detectable virus-specific molecules can be of viral origin, which could have a significant impact on the use of adenoviral vectors for gene therapy.
Figures

References
-
- Boyer J, Rohleder K, Ketner G. Adenovirus E4 34k and E4 11k inhibit double-strand-break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology. 1999;263:307–312. - PubMed
-
- Bradley W E, Letovanec D. High-frequency nonrandom mutational event at the adenine phosphoribosyltransferase (aprt) locus of sib-selected CHO variants heterozygous for aprt. Somatic Cell Genet. 1982;8:51–66. - PubMed
-
- Caporossi D, Bacchetti S. Definition of adenovirus type 5 functions involved in the induction of chromosomal aberrations in human cells. J Gen Virol. 1990;71:801–808. - PubMed
-
- Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. - PubMed
-
- Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

