Purification and characterization of West Nile virus nucleoside triphosphatase (NTPase)/helicase: evidence for dissociation of the NTPase and helicase activities of the enzyme
- PMID: 11238848
- PMCID: PMC114115
- DOI: 10.1128/JVI.75.7.3220-3229.2001
Purification and characterization of West Nile virus nucleoside triphosphatase (NTPase)/helicase: evidence for dissociation of the NTPase and helicase activities of the enzyme
Abstract
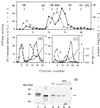
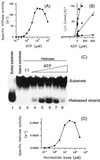
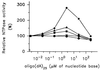
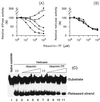
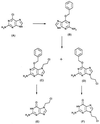
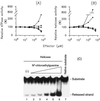
The nucleoside triphosphatase (NTPase)/helicase associated with nonstructural protein 3 of West Nile (WN) virus was purified from cell culture medium harvested from virus-infected Vero cells. The purification procedure included sequential chromatography on Superdex-200 and Reactive Red 120 columns, followed by a concentration step on an Ultrogel hydroxyapatite column. The nature of the purified protein was confirmed by immunoblot analysis using a WN virus-positive antiserum, determination of its NH(2) terminus by microsequencing, and a binding assay with 5'-[(14)C]fluorosulfonylbenzoyladenosine. Under optimized reaction conditions the enzyme catalyzed the hydrolysis of ATP and the unwinding of the DNA duplex with k(cat) values of 133 and 5.5 x 10(-3) s(-1), respectively. Characterization of the NTPase activity of the WN virus enzyme revealed that optimum conditions with respect to the Mg(2+) requirement and the monovalent salt or polynucleotide response differed from those of other flavivirus NTPases. Initial kinetic studies demonstrated that the inhibition (or activation) of ATPase activity by ribavirin-5'-triphosphate is not directly related to changes in the helicase activity of the enzyme. Further analysis using guanine and O(6)-benzoylguanine derivatives revealed that the ATPase activity of WN virus NTPase/helicase may be modulated, i.e., increased or reduced, with no effect on the helicase activity of the enzyme. On the other hand the helicase activity could be modulated without changing the ATPase activity. Our observations show that the number of ATP hydrolysis events per unwinding cycle is not a constant value.
Figures

References
-
- Ali J A, Lohman T M. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. - PubMed
-
- Bazan J F, Fletterick R. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989;171:637–639. - PubMed
-
- Borowski P, Heiland M, Oehlmann K, Becker B, Kornetzky L, Feucht H-H, Laufs R. Non-structural protein 3 of hepatitis C virus inhibits phosphorylation mediated by cAMP-dependent protein kinase. Eur J Biochem. 1996;237:611–618. - PubMed
-
- Borowski P, Kühl R, Laufs R, Schulze zur Wiesch J, Heiland M. Identification and characterization of a histone binding site of nonstructural protein 3 of hepatitis C virus. J Clin Virol. 1999;13:61–69. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

