Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity
- PMID: 11238853
- PMCID: PMC114120
- DOI: 10.1128/JVI.75.7.3268-3276.2001
Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity
Abstract
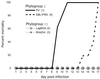
The genetic diversity of representative members of the Lyssavirus genus (rabies and rabies-related viruses) was evaluated using the gene encoding the transmembrane glycoprotein involved in the virus-host interaction, immunogenicity, and pathogenicity. Phylogenetic analysis distinguished seven genotypes, which could be divided into two major phylogroups having the highest bootstrap values. Phylogroup I comprises the worldwide genotype 1 (classic Rabies virus), the European bat lyssavirus (EBL) genotypes 5 (EBL1) and 6 (EBL2), the African genotype 4 (Duvenhage virus), and the Australian bat lyssavirus genotype 7. Phylogroup II comprises the divergent African genotypes 2 (Lagos bat virus) and 3 (Mokola virus). We studied immunogenic and pathogenic properties to investigate the biological significance of this phylogenetic grouping. Viruses from phylogroup I (Rabies virus and EBL1) were found to be pathogenic for mice when injected by the intracerebral or the intramuscular route, whereas viruses from phylogroup II (Mokola and Lagos bat viruses) were only pathogenic by the intracerebral route. We showed that the glycoprotein R333 residue essential for virulence was naturally replaced by a D333 in the phylogroup II viruses, likely resulting in their attenuated pathogenicity. Moreover, cross-neutralization distinguished the same phylogroups. Within each phylogroup, the amino acid sequence of the glycoprotein ectodomain was at least 74% identical, and antiglycoprotein virus-neutralizing antibodies displayed cross-neutralization. Between phylogroups, the identity was less than 64.5% and the cross-neutralization was absent, explaining why the classical rabies vaccines (phylogroup I) cannot protect against lyssaviruses from phylogroup II. Our tree-axial analysis divided lyssaviruses into two phylogroups that more closely reflect their biological characteristics than previous serotypes and genotypes.
Figures





References
-
- Amengual B, Whitby J E, King A, Cobo S, Bourhy H. Evolution of European bat lyssaviruses. J Gen Virol. 1997;78:2319–2328. - PubMed
-
- Atanasiu P, Tsiang H, Perrin P, Favre S. Demonstration of sialic acid in the rabies virus. Consequences of its removal on infectious and hemagglutinating properties. C R Acad Sc Hebd Seances Acad Sci D. 1976;283:111–114. - PubMed
-
- Bahloul C, Jacob Y, Tordo N, Perrin P. DNA-based immunisation for exploring the enlargement of immunological cross-reactivity against the lyssaviruses. Vaccine. 1998;16:417–425. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

