Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1
- PMID: 11238855
- PMCID: PMC114122
- DOI: 10.1128/JVI.75.7.3291-3300.2001
Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1
Abstract
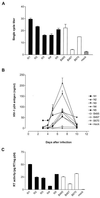
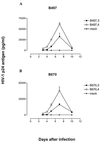
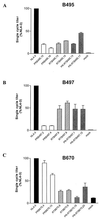
Human immunodeficiency virus type 1 (HIV-1) variants resistant to protease (PR) and reverse transcriptase (RT) inhibitors may display impaired infectivity and replication capacity. The individual contributions of mutated HIV-1 PR and RT to infectivity, replication, RT activity, and protein maturation (herein referred to as "fitness") in recombinant viruses were investigated by separately cloning PR, RT, and PR-RT cassettes from drug-resistant mutant viral isolates into the wild-type NL4-3 background. Both mutant PR and RT contributed to measurable deficits in fitness of viral constructs. In peripheral blood mononuclear cells, replication rates (means +/- standard deviations) of RT recombinants were 72.5% +/- 27.3% and replication rates of PR recombinants were 60.5% +/- 33.6% of the rates of NL4-3. PR mutant deficits were enhanced in CEM T cells, with relative replication rates of PR recombinants decreasing to 15.8% +/- 23.5% of NL4-3 replication rates. Cloning of the cognate RT improved fitness of some PR mutant clones. For a multidrug-resistant virus transmitted through sexual contact, RT constructs displayed a marked infectivity and replication deficit and diminished packaging of Pol proteins (RT content in virions diminished by 56.3% +/- 10.7%, and integrase content diminished by 23.3% +/- 18.4%), a novel mechanism for a decreased-fitness phenotype. Despite the identified impairment of recombinant clones, fitness of two of the three drug-resistant isolates was comparable to that of wild-type, susceptible viruses, suggestive of extensive compensation by genomic regions away from PR and RT. Only limited reversion of mutated positions to wild-type amino acids was observed for the native isolates over 100 viral replication cycles in the absence of drug selective pressure. These data underscore the complex relationship between PR and RT adaptive changes and viral evolution in antiretroviral drug-resistant HIV-1.
Figures



References
-
- Ansari-Lari A M, Donehower L A, Gibbs R A. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology. 1995;211:332–335. - PubMed
-
- Bahnson A. Centrifugal enhancement of retroviral mediated gene transfer. J Virol Methods. 1995;54:131–134. - PubMed
-
- Bally F, Martinez R, Peters S, Sudre P, Telenti A. Polymorphism of HIV-1 Gag p7/p1 and p1/p6 cleavage sites. Clinical significance and implications for resistance to protease inhibitors. AIDS Res Hum Retrovir. 2000;16:1209–1213. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

