Mitochondrial aconitase binds to the 3' untranslated region of the mouse hepatitis virus genome
- PMID: 11238861
- PMCID: PMC114128
- DOI: 10.1128/JVI.75.7.3352-3362.2001
Mitochondrial aconitase binds to the 3' untranslated region of the mouse hepatitis virus genome
Abstract
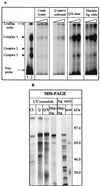
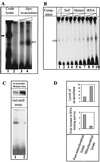
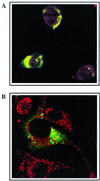
Mouse hepatitis virus (MHV), a member of the Coronaviridae, contains a polyadenylated positive-sense single-stranded genomic RNA which is 31 kb long. MHV replication and transcription take place via the synthesis of negative-strand RNA intermediates from a positive-strand genomic template. A cis-acting element previously identified in the 3' untranslated region binds to trans-acting host factors from mouse fibroblasts and forms at least three RNA-protein complexes. The largest RNA-protein complex formed by the cis-acting element and the lysate from uninfected mouse fibroblasts has a molecular weight of about 200 kDa. The complex observed in gel shift assays has been resolved by second-dimension sodium dodecyl sulfate-polyacrylamide gel electrophoresis into four proteins of approximately 90, 70, 58, and 40 kDa after RNase treatment. Specific RNA affinity chromatography also has revealed the presence of a 90-kDa protein associated with RNA containing the cis-acting element bound to magnetic beads. The 90-kDa protein has been purified from uninfected mouse fibroblast crude lysates. Protein microsequencing identified the 90-kDa protein as mitochondrial aconitase. Antibody raised against purified mitochondrial aconitase recognizes the RNA-protein complex and the 90-kDa protein, which can be released from the complex by RNase digestion. Furthermore, UV cross-linking studies indicate that highly purified mitochondrial aconitase binds specifically to the MHV 3' protein-binding element. Increasing the intracellular level of mitochondrial aconitase by iron supplementation resulted in increased RNA-binding activity in cell extracts and increased virus production as well as viral protein synthesis at early hours of infection. These results are particularly interesting in terms of identification of an RNA target for mitochondrial aconitase, which has a cytoplasmic homolog, cytoplasmic aconitase, also known as iron regulatory protein 1, a well-recognized RNA-binding protein. The binding properties of mitochondrial aconitase and the functional relevance of RNA binding appear to parallel those of cytoplasmic aconitase.
Figures



Similar articles
-
Viral and cellular proteins involved in coronavirus replication.Curr Top Microbiol Immunol. 2005;287:95-131. doi: 10.1007/3-540-26765-4_4. Curr Top Microbiol Immunol. 2005. PMID: 15609510 Free PMC article. Review.
-
Mitochondrial aconitase binds to the 3'-UTR of mouse hepatitis virus RNA.Adv Exp Med Biol. 2001;494:603-8. doi: 10.1007/978-1-4615-1325-4_89. Adv Exp Med Biol. 2001. PMID: 11774532 No abstract available.
-
A specific host cellular protein binding element near the 3' end of mouse hepatitis virus genomic RNA.Virology. 1997 May 26;232(1):74-85. doi: 10.1006/viro.1997.8553. Virology. 1997. PMID: 9185590
-
Effect of mutations in the mouse hepatitis virus 3'(+)42 protein binding element on RNA replication.J Virol. 2005 Dec;79(23):14570-85. doi: 10.1128/JVI.79.23.14570-14585.2005. J Virol. 2005. PMID: 16282457 Free PMC article.
-
Aconitase, a two-faced protein: enzyme and iron regulatory factor.FASEB J. 1993 Dec;7(15):1442-9. doi: 10.1096/fasebj.7.15.8262329. FASEB J. 1993. PMID: 8262329 Review.
Cited by
-
The structure and functions of coronavirus genomic 3' and 5' ends.Virus Res. 2015 Aug 3;206:120-33. doi: 10.1016/j.virusres.2015.02.025. Epub 2015 Feb 28. Virus Res. 2015. PMID: 25736566 Free PMC article. Review.
-
Viral and cellular proteins involved in coronavirus replication.Curr Top Microbiol Immunol. 2005;287:95-131. doi: 10.1007/3-540-26765-4_4. Curr Top Microbiol Immunol. 2005. PMID: 15609510 Free PMC article. Review.
-
RNA-RNA and RNA-protein interactions in coronavirus replication and transcription.RNA Biol. 2011 Mar-Apr;8(2):237-48. doi: 10.4161/rna.8.2.14991. Epub 2011 Mar 1. RNA Biol. 2011. PMID: 21378501 Free PMC article. Review.
-
Binding of the 5'-untranslated region of coronavirus RNA to zinc finger CCHC-type and RNA-binding motif 1 enhances viral replication and transcription.Nucleic Acids Res. 2012 Jun;40(11):5065-77. doi: 10.1093/nar/gks165. Epub 2012 Feb 22. Nucleic Acids Res. 2012. PMID: 22362731 Free PMC article.
-
The Natural History, Pathobiology, and Clinical Manifestations of SARS-CoV-2 Infections.J Neuroimmune Pharmacol. 2020 Sep;15(3):359-386. doi: 10.1007/s11481-020-09944-5. Epub 2020 Jul 21. J Neuroimmune Pharmacol. 2020. PMID: 32696264 Free PMC article. Review.
References
-
- Beinert H, Kennedy M C. Aconitase, a two-faced protein: enzyme and iron regulatory factor. FASEB J. 1993;7:1442–1449. - PubMed
-
- Berndt P, Hobohm U, Langen H. Reliable automatic protein identification from matrix-assisted laser desorption/ionization mass spectrometric peptide fingerprints. Electrophoresis. 1999;20:3521–3526. - PubMed
-
- Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. - PubMed
-
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

