A link between DNA methylation and epigenetic silencing in transgenic Volvox carteri
- PMID: 11238991
- PMCID: PMC29749
- DOI: 10.1093/nar/29.6.1261
A link between DNA methylation and epigenetic silencing in transgenic Volvox carteri
Abstract
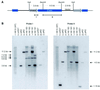
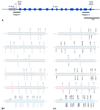
Epigenetic silencing of foreign genes introduced into plants poses an unsolved problem for transgenic technology. Here we have used the simple multicellular green alga VOLVOX: carteri as a model to analyse the relation of DNA methylation to transgenic silencing. VOLVOX: DNA contains on average 1.1% 5-methylcytosine and 0.3% N6-methyladenine, as revealed by electrospray mass spectrometry and phosphoimaging of chromatographically separated (32)P-labelled nucleotides. In two nuclear transformants of V.carteri, produced in 1993 by biolistic bombardment with a foreign arylsulphatase gene (C-ars), the transgene is still expressed in one (Hill 181), but not in the other (Hill 183), after an estimated 500-1000 generations. Each transformant clone contains multiple intact copies of C-ars, most of them integrated into the genome as tandem repeats. When the bisulphite genomic sequencing protocol was applied to examine two select regions of transgenic C-ars, we found that the inactivated copies (Hill 183) exhibited a high-level methylation (40%) of CpG dinucleotides, whereas the active copies (Hill 181) displayed low-level (7%) CpG methylation. These are average values from 40 PCR clones sequenced from each DNA strand in the two portions of C-ars. The observed correlation of CpG methylation and transgene inactivation in a green alga will be discussed in the light of transcriptional silencing.
Figures



References
-
- Schmitt R., Fabry,S. and Kirk,D.L. (1992) In search of molecular origins of cellular differentiation in Volvox and its relatives. Int. Rev. Cytol., 139, 189–265. - PubMed
-
- Kirk D.L. (1998) Volvox: Molecular Genetic Origins of Multicellularity and Cellular Differentiation. Cambridge University Press, Cambridge, UK.
-
- Kirk M.M., Stark,K., Miller,S.M., Müller,W., Taillon,B.E., Gruber,H., Schmitt,R. and Kirk,D.L. (1999) regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development, 126, 639–647. - PubMed
-
- Miller S.M. and Kirk,D.L. (1999) glsA, a Volvox gene required for asymmetric division and germ cell specification, encodes a chaperone-like protein. Development, 126, 649–658. - PubMed
-
- Meissner M., Stark,K., Cresnar,B., Kirk,D.L. and Schmitt,R. (1999) Volvox germline-specific genes that are putative targets of RegA repression encode chloroplast proteins. Curr. Genet., 36, 363–370. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

