Fully modified 2' MOE oligonucleotides redirect polyadenylation
- PMID: 11238995
- PMCID: PMC29745
- DOI: 10.1093/nar/29.6.1293
Fully modified 2' MOE oligonucleotides redirect polyadenylation
Abstract
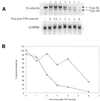
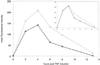
Many genes have been described and characterized that have alternative polyadenylation signals at the 3'-end of their pre-mRNAs. Many of these same messages also contain destabilization motifs responsible for rapid degradation of the mRNA. Polyadenylation site selection can thus determine the stability of an mRNA. Fully modified 2'-O:-methoxy ethyl/phosphorothioate oligonucleotides that hybridize to the 3'-most polyadenylation site or signal of E-selectin were able to inhibit polyadenylation at this site and redirect it to one of two upstream cryptic sites. The shorter transcripts produced after antisense treatment have fewer destabilization sequences, increased mRNA stability and altered protein expression. This study demonstrates that antisense oligonucleotides can be successfully employed to redirect polyadenylation. This is the first demonstration of the use of oligonucleotides to increase, rather than decrease, abundance of a message.
Figures



References
-
- Gautheret D., Poirot,O., Lopez,F., Audic,S. and Claverie,J.M. (1998) Alternate polyadenylation in human mRNAs: a large-scale analysis by EST clustering. Genome Res., 8, 524–530. - PubMed
-
- Shaw G. and Kamen,R. (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediated selective mRNA degradation. Cell, 46, 659–667. - PubMed
-
- Wahle E. and Keller,W. (1996) The biochemistry of polyadenylation. Trends Biochem. Sci., 21, 247–250. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

