UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals
- PMID: 11242562
- PMCID: PMC29092
- DOI: 10.1186/1471-2199-2-3
UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals
Abstract
Background: Translation termination is mediated through an interaction between the release factors eRF1 and eRF3 and the stop codon within its nucleotide context. Although it is well known that the nucleotide contexts both upstream and downstream of the stop codon, can modulate readthrough, little is known about the mechanisms involved.
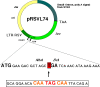
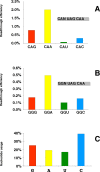
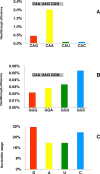
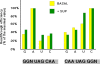
Results: We have performed an in vivo analysis of translational readthrough in mouse cells in culture using a reporter system that allows the measurement of readthrough levels as low as 10(-4). We first quantified readthrough frequencies obtained with constructs carrying different codons (two Gln, two His and four Gly) immediately upstream of the stop codon. There was no effect of amino acid identity or codon frequency. However, an adenine in the -1 position was always associated with the highest readthrough levels while an uracil was always associated with the lowest readthrough levels. This could be due to an effect mediated either by the nucleotide itself or by the P-site tRNA. We then examined the importance of the downstream context using eight other constructs. No direct correlation between the +6 nucleotide and readthrough efficiency was observed.
Conclusions: We conclude that, in mouse cells, the upstream and downstream stop codon contexts affect readthrough via different mechanisms, suggesting that complex interactions take place between the mRNA and the various components of the translation termination machinery. Comparison of our results with those previously obtained in plant cells and in yeast, strongly suggests that the mechanisms involved in stop codon recognition are conserved among eukaryotes.
Figures
References
-
- Kirkwood TBL, Rosenberger RF, Galas DJ. Accuracy in Molecular Processes. London: Chapman and Hall; 1986.
-
- Gesteland RF, Weiss RB, Atkins JF. Recoding: reprogrammed genetic decoding. Science. 1992;257:1640–1641. - PubMed
-
- Böck A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

