Genetic evidence that the alpha5 helix of the receiver domain of PhoB is involved in interdomain interactions
- PMID: 11244058
- PMCID: PMC95125
- DOI: 10.1128/JB.183.7.2204-2211.2001
Genetic evidence that the alpha5 helix of the receiver domain of PhoB is involved in interdomain interactions
Abstract
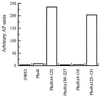
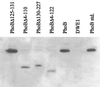
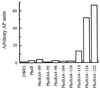
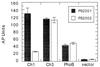
Two-component signaling proteins are involved in transducing environmental stimuli into intracellular signals. Information is transmitted through a phosphorylation cascade that consists of a histidine protein kinase and a response regulator protein. Generally, response regulators are made up of a receiver domain and an output domain. Phosphorylation of the receiver domain modulates the activity of the output domain. The mechanisms by which receiver domains control the activities of their respective output domains are unknown. To address this question for the PhoB protein from Escherichia coli, we have employed two separate genetic approaches, deletion analysis and domain swapping. In-frame deletions were generated within the phoB gene, and the phenotypes of the mutants were analyzed. The output domain, by itself, retained significant ability to activate transcription of the phoA gene. However, another deletion mutant that contained the C-terminal alpha-helix of the receiver domain (alpha5) in addition to the entire output domain was unable to activate transcription of phoA. This result suggests that the alpha5 helix of the receiver domain interacts with and inhibits the output domain. We also constructed two chimeric proteins that join various parts of the chemotaxis response regulator, CheY, to PhoB. A chimera that joins the N-terminal approximately 85% of CheY's receiver domain to the beta5-alpha5 loop of PhoB's receiver domain displayed phosphorylation-dependent activity. The results from both sets of experiments suggest that the regulation of PhoB involves the phosphorylation-mediated modulation of inhibitory contacts between the alpha5 helix of its unphosphorylated receiver domain and its output domain.
Figures


References
-
- Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. - PubMed
-
- Birck C, Mourey L, Gouet P, Fabry B, Schumacker J, Rousseau P, Kahn D, Samama J-P. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure. 1999;7:1505–1515. - PubMed
-
- Cho H S, Lee S Y, Yan D, Pan X, Parkinson J S, Kustu S, Wemmer D E, Pelton J G. NMR structure of activated CheY. J Mol Biol. 2000;297:543–551. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

