Aquaporins constitute a large and highly divergent protein family in maize
- PMID: 11244102
- PMCID: PMC65601
- DOI: 10.1104/pp.125.3.1206
Aquaporins constitute a large and highly divergent protein family in maize
Abstract
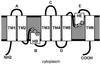
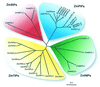
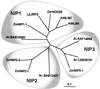
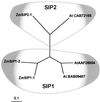
Aquaporins (AQPs) are an ancient family of channel proteins that transport water and neutral solutes through a pore and are found in all eukaryotes and most prokaryotes. A comparison of the amino acid sequences and phylogenetic analysis of 31 full-length cDNAs of maize (Zea mays) AQPs shows that they comprise four different groups of highly divergent proteins. We have classified them as plasma membrane intinsic proteins (PIPs), tonoplast intrinsic proteins, Nod26-like intrinsic proteins, and small and basic intrinsic proteins. Amino acid sequence identities vary from 16% to 100%, but all sequences share structural motifs and conserved amino acids necessary to stabilize the two loops that form the aqueous pore. Most divergent are the small and basic integral proteins in which the first of the two highly conserved Asn-Pro-Ala motifs of the pore is not conserved, but is represented by alanine-proline-threonine or alanine-proline-serine. We present a model of ZmPIP1-2 based on the three-dimensional structure of mammalian AQP1. Tabulation of the number of times that the AQP sequences are found in a collection of databases that comprises about 470,000 maize cDNAs indicates that a few of the maize AQPs are very highly expressed and many are not abundantly expressed. The phylogenetic analysis supports the interpretation that the divergence of PIPs through gene duplication occurred more recently than the divergence of the members of the other three subfamilies. This study opens the way to analyze the function of the proteins in Xenopus laevis oocytes, determine the tissue specific expression of the genes, recover insertion mutants, and determine the in planta function.
Figures



References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Barkla BJ, Vera-Estrella R, Pantoja O, Kirch HH, Bohnert HJ. Aquaporin localization: how valid are the TIP and PIP labels? Trends Plant Sci. 1999;4:86–88. - PubMed
-
- Biela A, Grote K, Otto B, Hoth S, Hedrich R, Kaldenhoff R. The Nicotiana tabacummembrane aquaporin NtAQP1 is mercury-insensitive and permeable for glycerol. Plant J. 1999;18:565–570. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

