Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene
- PMID: 11244106
- PMCID: PMC65605
- DOI: 10.1104/pp.125.3.1248
Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene
Abstract
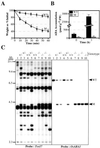
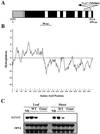
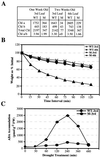
The rice (Oryza sativa) retrotransposon Tos17 is one of a few active retrotransposons in plants and its transposition is activated by tissue culture. Here, we present the characterization of viviparous mutants of rice induced by tissue culture to demonstrate the feasibility of the use of retrotransposon Tos17 as an endogenous insertional mutagen and cloning of the tagged gene for forward genetics in unraveling the gene function. Two mutants were shown to be caused by the insertion of Tos17. Osaba1, a strong viviparous mutant with wilty phenotype, displayed low abscisic acid level and almost no further increase in its levels upon drought. The mutant is shown to be impaired in the epoxidation of zeaxanthin. On the other hand, Ostatc, a mutant with weak phenotype, exhibited the pale green phenotype and slight increase in abscisic acid levels upon drought. Deduced amino acids of the causative genes of Osaba1 and Ostatc manifested a significantly high homology with zeaxanthin epoxidase isolated from other plant species and with bacterial Sec-independent translocase TATC protein, respectively. This is the first example of transposon tagging in rice.
Figures



References
-
- Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet. 1997;13:152–156. - PubMed
-
- Berks BC, Sargent F, Palmer T. The Tat protein export pathway. Mol Microbiol. 2000;35:260–274. - PubMed
-
- Bray EA. Plant responses to water deficit. Trends Plant Sci. 1997;2:48–54.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

