Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes
- PMID: 11244114
- PMCID: PMC65613
- DOI: 10.1104/pp.125.3.1342
Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes
Abstract
Colinearity of a large region from barley (Hordeum vulgare) chromosome 5H and rice (Oryza sativa) chromosome 3 has been demonstrated by mapping of several common restriction fragment-length polymorphism clones on both regions. One of these clones, WG644, was hybridized to rice and barley bacterial artificial chromosome (BAC) libraries to select homologous clones. One BAC from each species with the largest overlapping segment was selected by fingerprinting and blot hybridization with three additional restriction fragment-length polymorphism clones. The complete barley BAC 635P2 and a 50-kb segment of the rice BAC 36I5 were completely sequenced. A comparison of the rice and barley DNA sequences revealed the presence of four conserved regions, containing four predicted genes. The four genes are in the same orientation in rice, but the second gene is in inverted orientation in barley. The fourth gene is duplicated in tandem in barley but not in rice. Comparison of the homeologous barley and rice sequences assisted the gene identification process and helped determine individual gene structures. General gene structure (exon number, size, and location) was largely conserved between rice and barley and to a lesser extent with homologous genes in Arabidopsis. Colinearity of these four genes is not conserved in Arabidopsis compared with the two grass species. Extensive similarity was not found between the rice and barley sequences other than within the exons of the structural genes, and short stretches of homology in the promoters and 3' untranslated regions. The larger distances between the first three genes in barley compared with rice are explained by the insertion of different transposable retroelements.
Figures

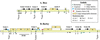

References
-
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218.
-
- Avramova Z, Tikhonov A, SanMiguel P, Jin YK, Liu CN, Woo SS, Wing RA, Bennetzen JL. Gene identification in a complex chromosomal continuum by local genomic cross-referencing. Plant J. 1996;10:1163–1168. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

