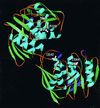
Closing down on glyphosate inhibition--with a new structure for drug discovery
- PMID: 11248008
- PMCID: PMC33334
- DOI: 10.1073/pnas.061025898
Closing down on glyphosate inhibition--with a new structure for drug discovery
Figures

Comment on
-
Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA.Proc Natl Acad Sci U S A. 2000 Jun 6;97(12):6345-9. doi: 10.1073/pnas.120120397. Proc Natl Acad Sci U S A. 2000. PMID: 10823915 Free PMC article.
-
Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.Proc Natl Acad Sci U S A. 2001 Feb 13;98(4):1376-80. doi: 10.1073/pnas.98.4.1376. Proc Natl Acad Sci U S A. 2001. PMID: 11171958 Free PMC article.
-
Structure and topological symmetry of the glyphosate target 5-enolpyruvylshikimate-3-phosphate synthase: a distinctive protein fold.Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):5046-50. doi: 10.1073/pnas.88.11.5046. Proc Natl Acad Sci U S A. 1991. PMID: 11607190 Free PMC article.
References
-
- Baird D D, Upchurch R P, Homesley W B, Franz J E. Proc North Cent Weed Control Conf. 1971;26:64–68.
-
- Franz J E, Mao M K, Sikorski J A. Glyphosate: A Unique Global Herbicide. Washington, DC: Am. Chem. Soc.; 1997.
-
- Jaworski E G. J Agric Food Chem. 1972;20:1195–1198.
-
- Haslam E. Shikimic Acid: Metabolism and Metabolites. New York: Wiley; 1993.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

