Structure of neurolysin reveals a deep channel that limits substrate access
- PMID: 11248043
- PMCID: PMC30618
- DOI: 10.1073/pnas.051633198
Structure of neurolysin reveals a deep channel that limits substrate access
Abstract
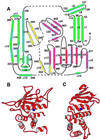
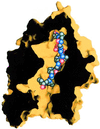
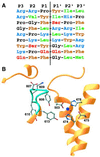
The zinc metallopeptidase neurolysin is shown by x-ray crystallography to have large structural elements erected over the active site region that allow substrate access only through a deep narrow channel. This architecture accounts for specialization of this neuropeptidase to small bioactive peptide substrates without bulky secondary and tertiary structures. In addition, modeling studies indicate that the length of a substrate N-terminal to the site of hydrolysis is restricted to approximately 10 residues by the limited size of the active site cavity. Some structural elements of neurolysin, including a five-stranded beta-sheet and the two active site helices, are conserved with other metallopeptidases. The connecting loop regions of these elements, however, are much extended in neurolysin, and they, together with other open coil elements, line the active site cavity. These potentially flexible elements may account for the ability of the enzyme to cleave a variety of sequences.
Figures
References
-
- Konkoy C S, Davis T P. Trends Pharmacol Sci. 1996;17:288–294. - PubMed
-
- Csuhai E, Safavi A, Thompson M W, Hersh L B. In: Proteolytic and Cellular Mechanisms in Prohormone and Neuropeptide Precursor Processing. Hook V, editor. Heidelberg: Springer; 1998. pp. 173–182.
-
- Checler F. In: Methods in Neurotransmitter and Neuropeptide Research. Nagatsu T, Parvez H, Naoi M, Parvez S, editors. Vol. 2. Amsterdam: Elsevier; 1993. pp. 375–418.
-
- Charli J L, Mendez M, Vargas M A, Cisneros M, Assai M, Joseph-Bravo P, Wilk S. Neuropeptides. 1989;14:191–196. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

