Cloning and mitochondrial localization of full-length D-AKAP2, a protein kinase A anchoring protein
- PMID: 11248059
- PMCID: PMC30634
- DOI: 10.1073/pnas.051633398
Cloning and mitochondrial localization of full-length D-AKAP2, a protein kinase A anchoring protein
Abstract
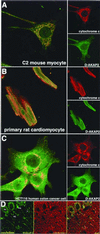
Differential compartmentalization of signaling molecules in cells and tissues is being recognized as an important mechanism for regulating the specificity of signal transduction pathways. A kinase anchoring proteins (AKAPs) direct the subcellular localization of protein kinase A (PKA) by binding to its regulatory (R) subunits. Dual specific AKAPs (D-AKAPs) interact with both RI and RII. A 372-residue fragment of mouse D-AKAP2 with a 40-residue C-terminal PKA binding region and a putative regulator of G protein signaling (RGS) domain was previously identified by means of a yeast two-hybrid screen. Here, we report the cloning of full-length human D-AKAP2 (662 residues) with an additional putative RGS domain, and the corresponding mouse protein less the first two exons (617 residues). Expression of D-AKAP2 was characterized by using mouse tissue extracts. Full-length D-AKAP2 from various tissues shows different molecular weights, possibly because of alternative splicing or posttranslational modifications. The cloned human gene product has a molecular weight similar to one of the prominent mouse proteins. In vivo association of D-AKAP2 with PKA in mouse brain was demonstrated by using cAMP agarose pull-down assay. Subcellular localization for endogenous mouse, rat, and human D-AKAP2 was determined by immunocytochemistry, immunohistochemistry, and tissue fractionation. D-AKAP2 from all three species is highly enriched in mitochondria. The mitochondrial localization and the presence of RGS domains in D-AKAP2 may have important implications for its function in PKA and G protein signal transduction.
Figures




References
-
- Cummings D E, Brandon E P, Planas J V, Motamed K, Idzerda R L, McKnight G S. Nature (London) 1996;382:622–626. - PubMed
-
- Jones P M, Sayed S B, Persaud S J, Burns C J, Gyles S, Whitehouse B J. J Mol Endocrinol. 2000;24:233–239. - PubMed
-
- Whitehouse B J, Abayasekara D R. J Mol Endocrinol. 1994;12:195–202. - PubMed
-
- Chen T C, Hinton D R, Zidovetzki R, Hofman F M. Lab Invest. 1998;78:165–174. - PubMed
-
- Tasken K, Skalhegg B S, Tasken K A, Solberg R, Knutsen H K, Levy F O, Sandberg M, Orstavik S, Larsen T, Johansen A K, et al. Adv Second Messenger Phosphoprotein Res. 1997;31:191–204. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

