Ligand-independent assembly of recombinant human CD1 by using oxidative refolding chromatography
- PMID: 11248071
- PMCID: PMC30646
- DOI: 10.1073/pnas.041596598
Ligand-independent assembly of recombinant human CD1 by using oxidative refolding chromatography
Abstract
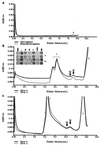
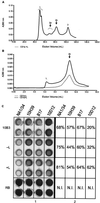
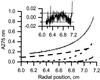
CD1 is an MHC class I-like antigen-presenting molecule consisting of a heavy chain and beta(2)-microglobulin light chain. The in vitro refolding of synthetic MHC class I molecules has always required the presence of ligand. We report here the use of a folding method using an immobilized chaperone fragment, a protein disulphide isomerase, and a peptidyl-prolyl cis-trans isomerase (oxidative refolding chromatography) for the fast and efficient assembly of ligand-free and ligand-associated CD1a and CD1b, starting with material synthesized in Escherichia coli. The results suggest that "empty" MHC class I-like molecules can assemble and remain stable at physiological temperatures in the absence of ligand. The use of oxidative refolding chromatography thus is extended to encompass complex multisubunit proteins and specifically to members of the extensive, functionally diverse and important immunoglobulin supergene family of proteins, including those for which a ligand has yet to be identified.
Figures


Comment in
-
Right on target: novel approaches for the direct visualization of CD1-specific T cell responses.Proc Natl Acad Sci U S A. 2001 Mar 13;98(6):2950-2. doi: 10.1073/pnas.061032398. Proc Natl Acad Sci U S A. 2001. PMID: 11248010 Free PMC article. Review. No abstract available.
References
-
- Calabi, F. and Milstein, C. (2001) Semin. Immunol., in press. - PubMed
-
- Porcelli S A, Modlin R L. Annu Rev Immunol. 1999;17:297–329. - PubMed
-
- Calabi F, Yung Yu C, Bilsland C A G, Milstein C. In: CD1: From Structure to Function. Immunogenetics of the Major Histocompatibility Complex. Srivastava R, Ram B, Tyle P, editors. NY: VCH; 1991.
-
- Burdin N, Kronenberg M. Curr Opin Immunol. 1999;3:326–331. - PubMed
-
- Balk S P, Burke S, Polischuk J E, Frantz M E, Yang L, Porcelli S, Colgan S P, Blumberg R S. Science. 1994;265:259–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

