The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal
- PMID: 11248102
- PMCID: PMC30677
- DOI: 10.1073/pnas.051484198
The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal
Abstract
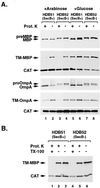
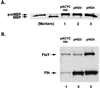
Previous studies have demonstrated that presecretory proteins such as maltose binding protein (MBP) and outer membrane protein A (OmpA) are targeted to the Escherichia coli inner membrane by the molecular chaperone SecB, but that integral membrane proteins are targeted by the signal recognition particle (SRP). In vitro studies have suggested that trigger factor binds to a sequence near the N terminus of the mature region of OmpA and shunts the protein into the SecB pathway by blocking an interaction between SRP and the signal peptide. By contrast, we have found that the targeting pathway of a protein under physiological conditions is dictated by the composition of its targeting signal. Replacement of the MBP or OmpA signal peptide with the first transmembrane segment of AcrB abolished the dependence on SecB for transport and rerouted both proteins into the SRP targeting pathway. More modest alterations of the MBP signal peptide that simply increase its hydrophobicity also promoted SRP binding. Furthermore, we obtained evidence that SRP has a low affinity for typical signal peptides in vivo. These results imply that different classes of E. coli proteins are targeted by distinct pathways because bacterial SRP binds to a more restricted range of targeting signals than its eukaryotic counterpart.
Figures




Similar articles
-
Escherichia coli signal peptides direct inefficient secretion of an outer membrane protein (OmpA) and periplasmic proteins (maltose-binding protein, ribose-binding protein, and alkaline phosphatase) in Bacillus subtilis.J Bacteriol. 1994 May;176(10):3013-20. doi: 10.1128/jb.176.10.3013-3020.1994. J Bacteriol. 1994. PMID: 8188602 Free PMC article.
-
SecA is required for the insertion of inner membrane proteins targeted by the Escherichia coli signal recognition particle.J Biol Chem. 1999 Mar 26;274(13):8993-7. doi: 10.1074/jbc.274.13.8993. J Biol Chem. 1999. PMID: 10085146
-
Export of maltose-binding protein species with altered charge distribution surrounding the signal peptide hydrophobic core in Escherichia coli cells harboring prl suppressor mutations.J Bacteriol. 1992 Jan;174(1):92-101. doi: 10.1128/jb.174.1.92-101.1992. J Bacteriol. 1992. PMID: 1729228 Free PMC article.
-
Export of the periplasmic maltose-binding protein of Escherichia coli.J Bioenerg Biomembr. 1990 Jun;22(3):401-39. doi: 10.1007/BF00763175. J Bioenerg Biomembr. 1990. PMID: 2202725 Review.
-
Mammalian and Escherichia coli signal recognition particles.Mol Microbiol. 1994 Jan;11(1):9-13. doi: 10.1111/j.1365-2958.1994.tb00284.x. Mol Microbiol. 1994. PMID: 8145649 Review.
Cited by
-
How Quality Control Systems AID Sec-Dependent Protein Translocation.Front Mol Biosci. 2021 Apr 13;8:669376. doi: 10.3389/fmolb.2021.669376. eCollection 2021. Front Mol Biosci. 2021. PMID: 33928127 Free PMC article. Review.
-
Prediction of signal recognition particle RNA genes.Nucleic Acids Res. 2002 Aug 1;30(15):3368-77. doi: 10.1093/nar/gkf468. Nucleic Acids Res. 2002. PMID: 12140321 Free PMC article.
-
Role for both DNA and RNA in GTP hydrolysis by the Neisseria gonorrhoeae signal recognition particle receptor.J Bacteriol. 2003 Feb;185(3):801-8. doi: 10.1128/JB.185.3.801-808.2003. J Bacteriol. 2003. PMID: 12533455 Free PMC article.
-
Pushing the Envelope: The Mysterious Journey Through the Bacterial Secretory Machinery, and Beyond.Front Microbiol. 2021 Nov 30;12:782900. doi: 10.3389/fmicb.2021.782900. eCollection 2021. Front Microbiol. 2021. PMID: 34917061 Free PMC article. Review.
-
Mutagenesis of DsbAss is Crucial for the Signal Recognition Particle Mechanism in Escherichia coli: Insights from Molecular Dynamics Simulations.Biomolecules. 2019 Apr 3;9(4):133. doi: 10.3390/biom9040133. Biomolecules. 2019. PMID: 30987187 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

