Non-templated addition of nucleotides to the 3' end of nascent RNA during RNA editing in Physarum
- PMID: 11250906
- PMCID: PMC145535
- DOI: 10.1093/emboj/20.6.1405
Non-templated addition of nucleotides to the 3' end of nascent RNA during RNA editing in Physarum
Abstract
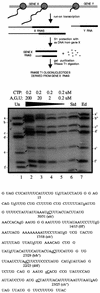
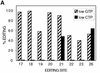
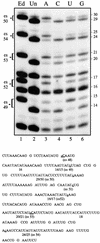
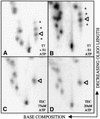
RNAs in Physarum: mitochondria contain extra nucleotides that are not encoded by the mitochondrial genome, at least in the traditional sense. While it is known that insertion of non-encoded nucleotides is linked to RNA synthesis, the exact nature of this relationship remains unclear. Here we demonstrate that the efficiency of editing is sensitive not only to the concentration of the nucleotide that is inserted, but also to the concentration of the nucleotide templated just downstream of an editing site. These data strongly support a co-transcriptional mechanism of Physarum: RNA editing in which non-encoded nucleotides are added to the 3' end of nascent RNAs. These results also suggest that transcription elongation and nucleotide insertion are competing processes and that recognition of editing sites most likely involves transient pausing by the Physarum: mitochondrial RNA polymerase. In addition, the pattern of nucleotide concentration effects, the context of editing sites and the accuracy of the mitochondrial RNA polymerase argue that the mechanism of Physarum: editing is distinct from that of other co-transcriptional editing systems.
Figures

Similar articles
-
Insertional editing in isolated Physarum mitochondria is linked to RNA synthesis.RNA. 1997 Aug;3(8):821-37. RNA. 1997. PMID: 9257642 Free PMC article.
-
Insertional editing of nascent mitochondrial RNAs in Physarum.Proc Natl Acad Sci U S A. 1997 Apr 29;94(9):4324-9. doi: 10.1073/pnas.94.9.4324. Proc Natl Acad Sci U S A. 1997. PMID: 9113988 Free PMC article.
-
Transcription and RNA editing in a soluble in vitro system from Physarum mitochondria.Nucleic Acids Res. 2000 Oct 1;28(19):3695-701. doi: 10.1093/nar/28.19.3695. Nucleic Acids Res. 2000. PMID: 11000260 Free PMC article.
-
Insertional editing in mitochondria of Physarum.Semin Cell Biol. 1993 Aug;4(4):261-6. doi: 10.1006/scel.1993.1031. Semin Cell Biol. 1993. PMID: 7694673 Review.
-
RNA editing.Annu Rev Neurosci. 1996;19:27-52. doi: 10.1146/annurev.ne.19.030196.000331. Annu Rev Neurosci. 1996. PMID: 8833435 Review.
Cited by
-
Unexpectedly complex editing patterns at dinucleotide insertion sites in Physarum mitochondria.Mol Cell Biol. 2004 Sep;24(18):7821-8. doi: 10.1128/MCB.24.18.7821-7828.2004. Mol Cell Biol. 2004. PMID: 15340046 Free PMC article.
-
Two forms of RNA editing are required for tRNA maturation in Physarum mitochondria.RNA. 2010 Mar;16(3):482-8. doi: 10.1261/rna.1958810. Epub 2010 Jan 27. RNA. 2010. PMID: 20106952 Free PMC article.
-
Identification of a putative mitochondrial RNA polymerase from Physarum polycephalum: characterization, expression, purification, and transcription in vitro.Curr Genet. 2006 Apr;49(4):259-71. doi: 10.1007/s00294-005-0053-y. Epub 2006 Jan 10. Curr Genet. 2006. PMID: 16402203
-
Discovery of new genes and deletion editing in Physarum mitochondria enabled by a novel algorithm for finding edited mRNAs.Nucleic Acids Res. 2005 Sep 7;33(16):5063-72. doi: 10.1093/nar/gki820. Print 2005. Nucleic Acids Res. 2005. PMID: 16147990 Free PMC article.
-
SCOPE++: sequence classification of homoPolymer emissions.Genomics. 2014 Sep;104(3):157-62. doi: 10.1016/j.ygeno.2014.07.005. Epub 2014 Aug 1. Genomics. 2014. PMID: 25087770 Free PMC article.
References
-
- Barik S., Ghosh,B., Whalen,W., Lazinski,D. and Das,A. (1987) An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell, 50, 885–899. - PubMed
-
- Bass B.L. (1993) RNA editing: new uses for old players in the RNA world. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 383–418.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

