Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA
- PMID: 11250910
- PMCID: PMC145536
- DOI: 10.1093/emboj/20.6.1449
Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA
Abstract
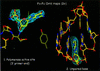
We have determined the 3.0 A resolution structure of wild-type HIV-1 reverse transcriptase in complex with an RNA:DNA oligonucleotide whose sequence includes a purine-rich segment from the HIV-1 genome called the polypurine tract (PPT). The PPT is resistant to ribonuclease H (RNase H) cleavage and is used as a primer for second DNA strand synthesis. The 'RNase H primer grip', consisting of amino acids that interact with the DNA primer strand, may contribute to RNase H catalysis and cleavage specificity. Cleavage specificity is also controlled by the width of the minor groove and the trajectory of the RNA:DNA, both of which are sequence dependent. An unusual 'unzipping' of 7 bp occurs in the adenine stretch of the PPT: an unpaired base on the template strand takes the base pairing out of register and then, following two offset base pairs, an unpaired base on the primer strand re-establishes the normal register. The structural aberration extends to the RNase H active site and may play a role in the resistance of PPT to RNase H cleavage.
Figures





Similar articles
-
Mechanism of polypurine tract primer generation by HIV-1 reverse transcriptase.J Biol Chem. 2018 Jan 5;293(1):191-202. doi: 10.1074/jbc.M117.798256. Epub 2017 Nov 9. J Biol Chem. 2018. PMID: 29122886 Free PMC article.
-
Two modes of HIV-1 polypurine tract cleavage are affected by introducing locked nucleic acid analogs into the (-) DNA template.J Biol Chem. 2004 Aug 27;279(35):37095-102. doi: 10.1074/jbc.M403306200. Epub 2004 Jun 25. J Biol Chem. 2004. PMID: 15220330
-
An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H.J Mol Biol. 2003 Dec 5;334(4):653-65. doi: 10.1016/j.jmb.2003.09.057. J Mol Biol. 2003. PMID: 14636594
-
'Binding, bending and bonding': polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus.Int J Biochem Cell Biol. 2004 Sep;36(9):1752-66. doi: 10.1016/j.biocel.2004.02.016. Int J Biochem Cell Biol. 2004. PMID: 15183342 Review.
-
Protein-nucleic acid interactions and DNA conformation in a complex of human immunodeficiency virus type 1 reverse transcriptase with a double-stranded DNA template-primer.Biopolymers. 1997;44(2):125-38. doi: 10.1002/(SICI)1097-0282(1997)44:2<125::AID-BIP2>3.0.CO;2-X. Biopolymers. 1997. PMID: 9354757 Review.
Cited by
-
Effects of HIV-1 reverse transcriptase connection subdomain mutations on polypurine tract removal and initiation of (+)-strand DNA synthesis.Nucleic Acids Res. 2015 Feb 27;43(4):2259-70. doi: 10.1093/nar/gkv077. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662223 Free PMC article.
-
Understanding the molecular mechanism of sequence dependent tenofovir removal by HIV-1 reverse transcriptase: differences in primer binding site versus polypurine tract.Antiviral Res. 2012 Aug;95(2):93-103. doi: 10.1016/j.antiviral.2012.05.012. Epub 2012 Jun 1. Antiviral Res. 2012. PMID: 22664235 Free PMC article.
-
Free Energy-Based Virtual Screening and Optimization of RNase H Inhibitors of HIV-1 Reverse Transcriptase.ACS Omega. 2016 Sep 30;1(3):435-447. doi: 10.1021/acsomega.6b00123. Epub 2016 Sep 21. ACS Omega. 2016. PMID: 27713931 Free PMC article.
-
TRIM5 and the Regulation of HIV-1 Infectivity.Mol Biol Int. 2012;2012:426840. doi: 10.1155/2012/426840. Epub 2012 May 30. Mol Biol Int. 2012. PMID: 22701176 Free PMC article.
-
Mn2+ suppressor mutations and biochemical communication between Ty1 reverse transcriptase and RNase H domains.J Virol. 2007 Sep;81(17):9004-12. doi: 10.1128/JVI.02502-06. Epub 2007 May 30. J Virol. 2007. PMID: 17537863 Free PMC article.
References
-
- Arnott S., Chandrasekaran,R., Millane,R.P. and Park,H.S. (1986) DNA-RNA hybrid secondary structures. J. Mol. Biol., 188, 631–640. - PubMed
-
- Bachelin M., Hessler,G., Kurz,G., Hacia,J.G., Dervan,P.B. and Kessler,H. (1998) Structure of a stereoregular phosphorothioate DNA/RNA duplex. Nature Struct. Biol., 5, 271–276. - PubMed
-
- Blain S.W. and Goff,S.P. (1993) Nuclease activities of Moloney murine leukemia virus reverse transcriptase. Mutants with altered substrate specificities. J. Biol. Chem., 268, 23585–23592. - PubMed
-
- Brünger A.T. (1993) X-PLOR Manual Version 3.1: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

