Expression and post-transcriptional regulation of maize transposable element MuDR and its derivatives
- PMID: 11251096
- PMCID: PMC135511
- DOI: 10.1105/tpc.13.3.553
Expression and post-transcriptional regulation of maize transposable element MuDR and its derivatives
Abstract
The transposition of Mu elements underlying Mutator activity in maize requires a transcriptionally active MuDR element. Despite variation in MuDR copy number and RNA levels in Mutator lines, transposition events are consistently late in plant development, and Mu excision frequencies are similar. Here, we report previously unsuspected and ubiquitous MuDR homologs that produce both RNA and protein. MuDR transcript levels are proportional to MuDR copy number, and homolog transcript levels increase in active Mutator lines. A subset of homologs exhibits constitutive transcription in MuDR(-) and epigenetically silenced MuDR lines, suggesting independent transcriptional regulation. Surprisingly, immunodetection demonstrated nearly invariant levels of MuDR and homolog protein products in all tested Mutator and non-Mutator stocks. These results suggest a strict control over protein production, which might explain the uniform excision frequency of Mu elements. Moreover, the nonfunctional proteins encoded by homologs may negatively regulate Mutator activity and represent part of the host defense against this transposon family.
Figures
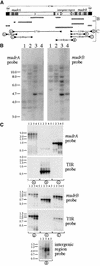
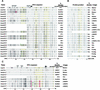

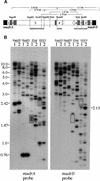

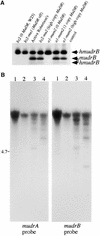

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

