Chemical genetics: ligand-based discovery of gene function
- PMID: 11253651
- PMCID: PMC3171795
- DOI: 10.1038/35038557
Chemical genetics: ligand-based discovery of gene function
Abstract
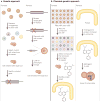
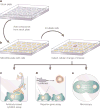
Chemical genetics is the study of gene-product function in a cellular or organismal context using exogenous ligands. In this approach, small molecules that bind directly to proteins are used to alter protein function, enabling a kinetic analysis of the in vivo consequences of these changes. Recent advances have strongly enhanced the power of exogenous ligands such that they can resemble genetic mutations in terms of their general applicability and target specificity. The growing sophistication of this approach raises the possibility of its application to any biological process.
Figures

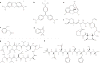
References
-
- Metzstein MM, Stanfield GM, Horvitz HR. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 1998;14:410–416. - PubMed
-
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. - PubMed
-
- Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. Mouse ENU mutagenesis. Hum Mol Genet. 1999;8:1955–1963. - PubMed
-
- Mullins MC, Hammerschmidt M, Haffter P, Nusslein-Volhard C. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr Biol. 1994;4:189–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

