An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations
- PMID: 11254455
- PMCID: PMC1275652
- DOI: 10.1086/319510
An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations
Abstract
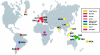
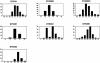
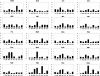
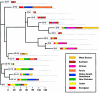
The genetic variance at seven Y-chromosomal microsatellite loci (or short tandem repeats [STRs]) was studied among 986 male individuals from 20 globally dispersed human populations. A total of 598 different haplotypes were observed, of which 437 (73.1%) were each found in a single male only. Population-specific haplotype-diversity values were.86-.99. Analyses of haplotype diversity and population-specific haplotypes revealed marked population-structure differences between more-isolated indigenous populations (e.g., Central African Pygmies or Greenland Inuit) and more-admixed populations (e.g., Europeans or Surinamese). Furthermore, male individuals from isolated indigenous populations shared haplotypes mainly with male individuals from their own population. By analysis of molecular variance, we found that 76.8% of the total genetic variance present among these male individuals could be attributed to genetic differences between male individuals who were members of the same population. Haplotype sharing between populations, phi(ST) statistics, and phylogenetic analysis identified close genetic affinities among European populations and among New Guinean populations. Our data illustrate that Y-chromosomal STR haplotypes are an ideal tool for the study of the genetic affinities between groups of male subjects and for detection of population structure.
Figures


References
Electronic-Database Information
-
- ARLEQUIN: A Software For Population Genetic Data Analysis, http://anthropologie.unige.ch/arlequin
-
- Rod Page's Home Page, http://taxonomy.zoology.gla.ac.uk/rod/rod.html (for TREEVIEW 1.6.1)
-
- Forensic Laboratory for DNA Research, http://www.medfac.leidenuniv.nl/fldo (for marker information)
-
- Y-STR Haplotype Reference Database, http://ystr.charite.de/index_gr.html (for marker information)
References
-
- Armour JA, Anttinen T, May CA, Vega EE, Sajantila A, Kidd JR, Kidd KK, Bertranpetit J, Pääbo S, Jeffreys AJ (1996) Minisatellite diversity supports a recent African origin for modern humans. Nat Genet 13:154–160 - PubMed
-
- Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

