High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula
- PMID: 11254456
- PMCID: PMC1275654
- DOI: 10.1086/319521
High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula
Abstract
In the present study we have analyzed 44 Y-chromosome biallelic polymorphisms in population samples from northwestern (NW) Africa and the Iberian Peninsula, which allowed us to place each chromosome unequivocally in a phylogenetic tree based on >150 polymorphisms. The most striking results are that contemporary NW African and Iberian populations were found to have originated from distinctly different patrilineages and that the Strait of Gibraltar seems to have acted as a strong (although not complete) barrier to gene flow. In NW African populations, an Upper Paleolithic colonization that probably had its origin in eastern Africa contributed 75% of the current gene pool. In comparison, approximately 78% of contemporary Iberian Y chromosomes originated in an Upper Paleolithic expansion from western Asia, along the northern rim of the Mediterranean basin. Smaller contributions to these gene pools (constituting 13% of Y chromosomes in NW Africa and 10% of Y chromosomes in Iberia) came from the Middle East during the Neolithic and, during subsequent gene flow, from Sub-Saharan to NW Africa. Finally, bidirectional gene flow across the Strait of Gibraltar has been detected: the genetic contribution of European Y chromosomes to the NW African gene pool is estimated at 4%, and NW African populations may have contributed 7% of Iberian Y chromosomes. The Islamic rule of Spain, which began in a.d. 711 and lasted almost 8 centuries, left only a minor contribution to the current Iberian Y-chromosome pool. The high-resolution analysis of the Y chromosome allows us to separate successive migratory components and to precisely quantify each historical layer.
Figures
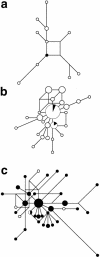
Comment in
-
Genetic evidence for the expansion of Arabian tribes into the Southern Levant and North Africa.Am J Hum Genet. 2002 Jun;70(6):1594-6. doi: 10.1086/340669. Am J Hum Genet. 2002. PMID: 11992266 Free PMC article. No abstract available.
References
Electronic-Database Information
-
- Genetree, http://www.maths.monash.edu.au/~mbahlo/mpg/gtree.html (software for coalescence analysis of gene genealogies)
-
- www.fluxus-engineering.com, http://www.fluxus-engineering.com (for Network 2.0c software for median-joining network construction)
References
-
- Bahlo M, Griffiths RC (2000) Inference from gene trees in a subdivided population. Theor Popul Biol 57:79–95 - PubMed
-
- Bertranpetit J, Sala J, Calafell F, Underhill P, Moral P, Comas D (1995) Human mitochondrial DNA variation and the origin of the Basques. Ann Hum Genet 59:63–81 - PubMed
-
- Bosch E, Calafell F, Pérez-Lezaun A, Clarimón J, Comas D, Mateu E, Martínez-Arias R, Morera B, Brakez Z, Akhayat O, Sefiani A, Harit G, Cambon-Thomsen A, Bertranpetit J (2000) Genetic structure of north-west Africa revealed by STR analysis. Eur J Hum Genet 8:360–366 - PubMed
-
- Bosch E, Calafell F, Pérez-Lezaun A, Comas D, Mateu E, Bertranpetit J (1997) A population history of northern Africa: evidence from classical genetic markers. Hum Biol 69:295–311 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

