Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis
- PMID: 11254558
- PMCID: PMC98130
- DOI: 10.1128/IAI.69.4.2054-2065.2001
Identification of new genes involved in the virulence of Listeria monocytogenes by signature-tagged transposon mutagenesis
Abstract
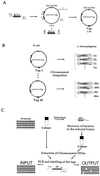
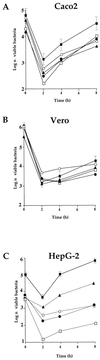
Listeria monocytogenes is a gram-positive, facultative intracellular pathogen that can cause severe food-born infections in humans and animals. We have adapted signature-tagged transposon mutagenesis to L. monocytogenes to identify new genes involved in virulence in the murine model of infection. We used transposon Tn1545 carried on the integrative vector pAT113. Forty-eight tagged transposons were constructed and used to generate banks of L. monocytogenes mutants. Pools of 48 mutants were assembled, taking one mutant from each bank, injected into mice, and screened for those affected in their multiplication in the brains of infected animals. From 2,000 mutants tested, 18 were attenuated in vivo. The insertions harbored by these mutants led to the identification of 10 distinct loci, 7 of which corresponded to previously unknown genes. The properties of four loci involving putative cell wall components were further studied in vitro and in vivo. The data suggested that these components are involved in bacterial invasion and multiplication in the brain.
Figures

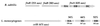



References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1990.
-
- Berche P. Bacteremia is required for invasion of the murine central nervous system by Listeria monocytogenes. Microb Pathog. 1995;18:323–336. - PubMed
-
- Berche P, Gaillard J L, Sansonetti P J. Intracellular growth of Listeria monocytogenes as a prerequisite for in vivo induction of T cell-mediated immunity. J Immunol. 1987;138:2266–2271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

