Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation of Streptococcus pneumoniae
- PMID: 11254610
- PMCID: PMC98182
- DOI: 10.1128/IAI.69.4.2477-2486.2001
Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation of Streptococcus pneumoniae
Abstract
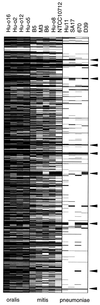
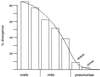
Streptococcus pneumoniae remains a major causative agent of serious human diseases. The worldwide increase of antibiotic resistant strains revealed the importance of horizontal gene transfer in this pathogen, a scenario that results in the modulation of the species-specific gene pool. We investigated genomic variation in 20 S. pneumoniae isolates representing major antibiotic-resistant clones and 10 different capsular serotypes. Variation was scored as decreased hybridization signals visualized on a high-density oligonucleotide array representing 1,968 genes of the type 4 reference strain KNR.7/87. Up to 10% of the genes appeared altered between individual isolates and the reference strain; variability within clones was below 2.1%. Ten gene clusters covering 160 kb account for half of the variable genes. Most of them are associated with transposases and are assumed to be part of a flexible gene pool within the bacterial population; other variable loci include mosaic genes encoding antibiotic resistance determinants and gene clusters related to bacteriocin production. Genomic comparison between S. pneumoniae and commensal Streptococcus mitis and Streptococcus oralis strains indicates distinct antigenic profiles and suggests a smooth transition between these species, supporting the validity of the microarray system as an epidemiological and diagnostic tool.
Figures





References
-
- Aaberge I S, Eng J, Lermark G, Lovik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb Pathog. 1995;18:141–152. - PubMed
-
- Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noona B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. - PubMed
-
- Baltz R H, Norris F H, Matsushima P, DeHoff B S, Rockey P, Porter G, Burgett S, Peery R, Hoskins J, Braverman L, Jenkins I, Solenburg P, Young M, McHenney M A, Skatrud P L, Rosteck P R., Jr DNA sequence sampling of the Streptococcus pneumoniae genome to identify novel targets for antibiotic development. Microb Drug Resist. 1998;4:1–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

