Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing
- PMID: 11256614
- PMCID: PMC1083732
- DOI: 10.1093/embo-reports/kvd058
Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing
Abstract
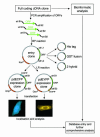
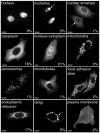
As a first step towards a more comprehensive functional characterization of cDNAs than bioinformatic analysis, which can only make functional predictions for about half of the cDNAs sequenced, we have developed and tested a strategy that allows their systematic and fast subcellular localization. We have used a novel cloning technology to rapidly generate N- and C-terminal green fluorescent protein fusions of cDNAs to examine the intracellular localizations of > 100 expressed fusion proteins in living cells. The entire analysis is suitable for automation, which will be important for scaling up throughput. For > 80% of these new proteins a clear intracellular localization to known structures or organelles could be determined. For the cDNAs where bioinformatic analyses were able to predict possible identities, the localization was able to support these predictions in 75% of cases. For those cDNAs where no homologies could be predicted, the localization data represent the first information.
Figures
References
-
- Bernard P., Gabant, P., Bahassi, E.M. and Couturier, M. (1994) Positive-selection vectors using the F plasmid ccdB killer gene. Gene, 148, 71–74. - PubMed
-
- Dunham I. et al. (1999) The DNA sequence of human chromosome 22. Nature, 402, 489–495. - PubMed
-
- Eisenhaber F. and Bork, P. (1998) Wanted: subcellular localization of proteins based on sequence. Trends Cell Biol., 8, 169–170. - PubMed
-
- Gonzalez C. and Bejarano, L.A. (2000) Protein traps: using intracellular localization for cloning. Trends Cell Biol., 10, 162–165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

