UGO1 encodes an outer membrane protein required for mitochondrial fusion
- PMID: 11257114
- PMCID: PMC2199209
- DOI: 10.1083/jcb.152.6.1123
UGO1 encodes an outer membrane protein required for mitochondrial fusion
Erratum in
- J Cell Biol 2001 Apr 30;153(3):635
Abstract
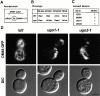
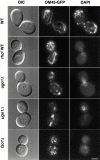
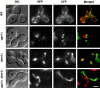
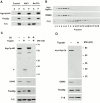
Membrane fusion plays an important role in controlling the shape, number, and distribution of mitochondria. In the yeast Saccharomyces cerevisiae, the outer membrane protein Fzo1p has been shown to mediate mitochondrial fusion. Using a novel genetic screen, we have isolated new mutants defective in the fusion of their mitochondria. One of these mutants, ugo1, shows several similarities to fzo1 mutants. ugo1 cells contain numerous mitochondrial fragments instead of the few long, tubular organelles seen in wild-type cells. ugo1 mutants lose mitochondrial DNA (mtDNA). In zygotes formed by mating two ugo1 cells, mitochondria do not fuse and mix their matrix contents. Fragmentation of mitochondria and loss of mtDNA in ugo1 mutants are rescued by disrupting DNM1, a gene required for mitochondrial division. We find that UGO1 encodes a 58-kD protein located in the mitochondrial outer membrane. Ugo1p appears to contain a single transmembrane segment, with its NH(2) terminus facing the cytosol and its COOH terminus in the intermembrane space. Our results suggest that Ugo1p is a new outer membrane component of the mitochondrial fusion machinery.
Figures




Similar articles
-
Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane.Mol Biol Cell. 2003 Jun;14(6):2342-56. doi: 10.1091/mbc.e02-12-0788. Epub 2003 Feb 6. Mol Biol Cell. 2003. PMID: 12808034 Free PMC article.
-
Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape.J Cell Biol. 1999 Nov 15;147(4):699-706. doi: 10.1083/jcb.147.4.699. J Cell Biol. 1999. PMID: 10562274 Free PMC article.
-
Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion.J Biol Chem. 2004 Jul 2;279(27):28298-303. doi: 10.1074/jbc.M401363200. Epub 2004 Apr 14. J Biol Chem. 2004. PMID: 15087460
-
Mitochondrial dynamics in yeast.Annu Rev Cell Dev Biol. 1998;14:265-303. doi: 10.1146/annurev.cellbio.14.1.265. Annu Rev Cell Dev Biol. 1998. PMID: 9891785 Review.
-
Merging mitochondria matters: cellular role and molecular machinery of mitochondrial fusion.EMBO Rep. 2002 Jun;3(6):527-31. doi: 10.1093/embo-reports/kvf113. EMBO Rep. 2002. PMID: 12052774 Free PMC article. Review.
Cited by
-
Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids.J Cell Biol. 2004 Mar 1;164(5):677-88. doi: 10.1083/jcb.200308012. Epub 2004 Feb 23. J Cell Biol. 2004. PMID: 14981098 Free PMC article.
-
A Molecular Perspective on Mitochondrial Membrane Fusion: From the Key Players to Oligomerization and Tethering of Mitofusin.J Membr Biol. 2019 Oct;252(4-5):293-306. doi: 10.1007/s00232-019-00089-y. Epub 2019 Sep 4. J Membr Biol. 2019. PMID: 31485701 Review.
-
Mitochondrial Stasis Reveals p62-Mediated Ubiquitination in Parkin-Independent Mitophagy and Mitigates Nonalcoholic Fatty Liver Disease.Cell Metab. 2018 Oct 2;28(4):588-604.e5. doi: 10.1016/j.cmet.2018.06.014. Epub 2018 Jul 12. Cell Metab. 2018. PMID: 30017357 Free PMC article.
-
The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion.J Cell Biol. 2003 Feb 3;160(3):303-11. doi: 10.1083/jcb.200209015. J Cell Biol. 2003. PMID: 12566426 Free PMC article.
-
The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins.J Cell Biol. 2011 Aug 8;194(3):387-95. doi: 10.1083/jcb.201102044. J Cell Biol. 2011. PMID: 21825073 Free PMC article.
References
-
- Adams A., Gottschling D., Kaiser C., Stearns T. Methods in Yeast Genetics 1997. Cold Spring Harbor Laboratory Press, ; Plainview, NY: pp. 177 pp
-
- Adams A.E., Pringle J.R. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 1991;194:729–731. - PubMed
-
- Bereiter-Hahn J., Voth M. Dynamics of mitochondria in living cellsshape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 1994;27:198–219. - PubMed
-
- Boeke J.D., LaCroute F., Fink G.R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast5-fluoro-orotic acid resistance. Mol. Gen. Genet. 1984;197:345–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

