Finding nuclear localization signals
- PMID: 11258480
- PMCID: PMC1083765
- DOI: 10.1093/embo-reports/kvd092
Finding nuclear localization signals
Abstract
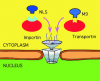
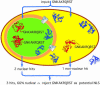
A variety of nuclear localization signals (NLSs) are experimentally known although only one motif was available for database searches through PROSITE. We initially collected a set of 91 experimentally verified NLSs from the literature. Through iterated 'in silico mutagenesis' we then extended the set to 214 potential NLSs. This final set matched in 43% of all known nuclear proteins and in no known non-nuclear protein. We estimated that >17% of all eukaryotic proteins may be imported into the nucleus. Finally, we found an overlap between the NLS and DNA-binding region for 90% of the proteins for which both the NLS and DNA-binding regions were known. Thus, evolution seemed to have used part of the existing DNA-binding mechanism when compartmentalizing DNA-binding proteins into the nucleus. However, only 56 of our 214 NLS motifs overlapped with DNA-binding regions. These 56 NLSs enabled a de novo prediction of partial DNA-binding regions for approximately 800 proteins in human, fly, worm and yeast.
Figures

References
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

