Molecular and cytological analysis of a 5.5 Mb minichromosome
- PMID: 11258700
- PMCID: PMC1083814
- DOI: 10.1093/embo-reports/kve018
Molecular and cytological analysis of a 5.5 Mb minichromosome
Abstract
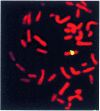
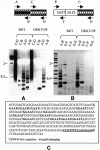
Mammalian artificial chromosomes (MACs) provide a new tool for the improvement of our knowledge of chromosome structure and function. Moreover, they constitute an alternative and potentially powerful tool for gene delivery both in cultured cells and for the production of transgenic animals. In the present work we describe the molecular structure of MC1, a human minichromosome derived from chromosome 1. By means of restriction and hybridization analysis, satellite-PCR, in situ hybridization on highly extended chromatin fibres, and indirect immunofluorescence, we have established that: (i) MC1 has a size of 5.5 Mb; (ii) it consists of 1.1 Mb alphoid, 3.5 Mb Sat2 DNA, and telomeric and subtelomeric sequences at both ends; (iii) it contains an unusual region of interspersed Sat2 and alphoid DNAs at the junction of the alphoid and the Sat2 blocks; and (iv) the two alphoid blocks and the Sat2-alphoid region bind centromeric proteins suggesting that they participate in the formation of a functional kinetochore.
Figures
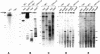


References
-
- Aleksandrov I.A., Akopian, T.A., Vinnik, E.A., Mitkevich, S.P. and Kiselev, L.L. (1988) Primary structure of chromosome-specific human alpha-satellite DNA. Dokl. Akad. Nauk SSSR, 300, 1235–1239. - PubMed
-
- Ascenzioni F., Della Valle, G., Guerrini, A.M., Pisani, G., Biondi, O. and Donini, P. (1990) A human DNA telomeric fragment containing yeast ARS and mitotic stabilizing sequences. Res. Microbiol., 141, 1117–1129. - PubMed
-
- Au H.C., Mascarello, J.T. and Scheffler, I.E. (1999) Targeted integration of a dominant neoR marker into a 2- to 3-Mb Human minichromosome and transfer between cells. Cytogenet. Cell Genet., 86, 194–203. - PubMed
-
- Bertoni L., Attolini, C., Faravelli, M., Simi, S. and Giulotto, E. (1996) Intrachromosomal telomere-like DNA sequences in Chinese hamster. Mamm. Genome, 7, 853–855. - PubMed
-
- Carine K., Solus, J., Waltzer, E., Manch-Citron, J., Hamkalo, B.A. and Scheffler, I.E. (1986) Chinese hamster cells with a minichromosome containing the centromeric region of human chromosome 1. Somat. Cell Mol. Genet., 12, 479–491. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

