The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition
- PMID: 11258702
- PMCID: PMC1083818
- DOI: 10.1093/embo-reports/kve022
The histone H4 acetyltransferase MOF uses a C2HC zinc finger for substrate recognition
Abstract
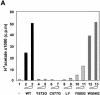
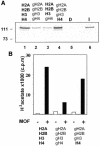
Site-specific acetylation of histone H4 by MOF is central to establishing the hyperactive male X chromosome in Drosophila. MOF belongs to the MYST family of histone acetyltransferases (HATs) characterized by an unusual C2HC-type zinc finger close to their HAT domains. The function of these rare zinc fingers is unknown. We found that this domain is essential for HAT activity, in addition to the established catalytic domain. MOF uses its zinc finger to contact the globular part of the nucleosome as well as the histone H4 N-terminal tail substrate. Point mutations that leave the zinc-finger structure intact nevertheless abolish its interaction with the nucleosome. Our data document a novel role of the C2HC-type finger in nucleosome binding and HAT activity.
Figures





References
-
- Akhtar A. and Becker, P.B. (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyl transferase essential for dosage compensation in Drosophila. Mol. Cell, 5, 367–375. - PubMed
-
- Akhtar A., Zink, D. and Becker, P.B. (2000) Chromodomains as RNA interaction modules. Nature, 407, 405–409. - PubMed
-
- Baker B.S., Gorman, M. and Marin, I. (1994) Dosage compensation in Drosophila. Annu. Rev. Genet., 28, 491–521. - PubMed
-
- Brown C.E., Lechner, T., Howe, L. and Workman, J.L. (2000). The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

