Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome
- PMID: 11259578
- PMCID: PMC86862
- DOI: 10.1128/MCB.21.7.2281-2291.2001
Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome
Abstract
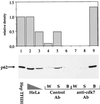
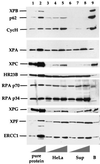
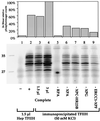
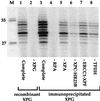
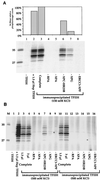
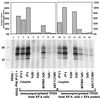
In mammalian cells, the core factors involved in the damage recognition and incision steps of DNA nucleotide excision repair are XPA, TFIIH complex, XPC-HR23B, replication protein A (RPA), XPG, and ERCC1-XPF. Many interactions between these components have been detected, using different physical methods, in human cells and for the homologous factors in Saccharomyces cerevisiae. Several human nucleotide excision repair (NER) complexes, including a high-molecular-mass repairosome complex, have been proposed. However, there have been no measurements of activity of any mammalian NER protein complex isolated under native conditions. In order to assess relative strengths of interactions between NER factors, we captured TFIIH from cell extracts with an anti-cdk7 antibody, retaining TFIIH in active form attached to magnetic beads. Coimmunoprecipitation of other NER proteins was then monitored functionally in a reconstituted repair system with purified proteins. We found that all detectable TFIIH in gently prepared human cell extracts was present in the intact nine-subunit form. There was no evidence for a repair complex that contained all of the NER components. At low ionic strength TFIIH could associate with functional amounts of each NER factor except RPA. At physiological ionic strength, TFIIH associated with significant amounts of XPC-HR23B and XPG but not other repair factors. The strongest interaction was between TFIIH and XPC-HR23B, indicating a coupled role of these proteins in early steps of repair. A panel of antibodies was used to estimate that there are on the order of 10(5) molecules of each core NER factor per HeLa cell.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
