Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family
- PMID: 11259589
- PMCID: PMC86873
- DOI: 10.1128/MCB.21.7.2404-2412.2001
Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family
Abstract
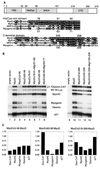
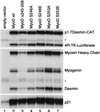
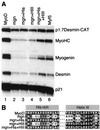
The myogenic basic helix-loop-helix (bHLH) proteins regulate both skeletal muscle specification and differentiation: MyoD and Myf5 establish the muscle lineage, whereas myogenin mediates differentiation. Previously, we demonstrated that MyoD was more efficient than myogenin at initiating the expression of skeletal muscle genes, and in this study we present the molecular basis for this difference. A conserved amphipathic alpha-helix in the carboxy terminus of the myogenic bHLH proteins has distinct activities in MyoD and myogenin: the MyoD helix facilitates the initiation of endogenous gene expression, whereas the myogenin helix functions as a general transcriptional activation domain. Thus, the alternate use of a similar motif for gene initiation and activation provides a molecular basis for the distinction between specification and differentiation within the myogenic bHLH gene family.
Figures




References
-
- Edmondson D G, Olson E N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989;3:628–640. . (Erratum, 4:1450, 1990.) - PubMed
-
- Fujisawa-Sehara A, Nabeshima Y, Hosoda Y, Obinata T. Myogenin contains two domains conserved among myogenic factors. J Biol Chem. 1990;265:15219–15223. - PubMed
-
- Gaudreau L, Schmid A, Blaschke D, Ptashne M, Horz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. - PubMed
-
- Gerber A N, Klesert T R, Bergstrom D A, Tapscott S J. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
