The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae
- PMID: 11259593
- PMCID: PMC86877
- DOI: 10.1128/MCB.21.7.2449-2462.2001
The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae
Abstract
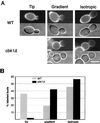
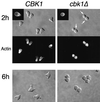
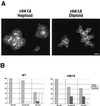
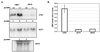
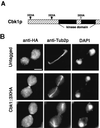
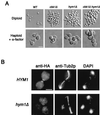
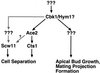
During the early stages of budding, cell wall remodeling and polarized secretion are concentrated at the bud tip (apical growth). The CBK1 gene, encoding a putative serine/threonine protein kinase, was identified in a screen designed to isolate mutations that affect apical growth. Analysis of cbk1Delta cells reveals that Cbk1p is required for efficient apical growth, proper mating projection morphology, bipolar bud site selection in diploid cells, and cell separation. Epitope-tagged Cbk1p localizes to both sides of the bud neck in late anaphase, just prior to cell separation. CBK1 and another gene, HYM1, were previously identified in a screen for genes involved in transcriptional repression and proposed to function in the same pathway. Deletion of HYM1 causes phenotypes similar to those observed in cbk1Delta cells and disrupts the bud neck localization of Cbk1p. Whole-genome transcriptional analysis of cbk1Delta suggests that the kinase regulates the expression of a number of genes with cell wall-related functions, including two genes required for efficient cell separation: the chitinase-encoding gene CTS1 and the glucanase-encoding gene SCW11. The Ace2p transcription factor is required for expression of CTS1 and has been shown to physically interact with Cbk1p. Analysis of ace2Delta cells reveals that Ace2p is required for cell separation but not for polarized growth. Our results suggest that Cbk1p and Hym1p function to regulate two distinct cell morphogenesis pathways: an ACE2-independent pathway that is required for efficient apical growth and mating projection formation and an ACE2-dependent pathway that is required for efficient cell separation following cytokinesis. Cbk1p is most closely related to the Neurospora crassa Cot-1; Schizosaccharomyces pombe Orb6; Caenorhabditis elegans, Drosophila, and human Ndr; and Drosophila and mammalian WARTS/LATS kinases. Many Cbk1-related kinases have been shown to regulate cellular morphology.
Figures






References
-
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 2000.
-
- Bedinger P A, Hardeman K J, Loukides C A. Travelling in style: the cell biology of pollen. Trends Cell Biol. 1994;4:132–138. - PubMed
-
- Belham C, Wu S, Avruch J. Intracellular signalling: PDK1—a kinase at the hub of things. Curr Biol. 1999;9:R93–R96. - PubMed
-
- Bulawa C E, Slater M, Cabib E, Au-Young J, Sburlati A, Adair W L, Jr, Robbins P W. The S. cerevisiae structural gene for chitin synthase is not required for chitin synthesis in vivo. Cell. 1986;46:213–225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
