Are all DNA binding and transcription regulation by an activator physiologically relevant?
- PMID: 11259595
- PMCID: PMC86879
- DOI: 10.1128/MCB.21.7.2467-2474.2001
Are all DNA binding and transcription regulation by an activator physiologically relevant?
Abstract
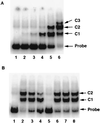
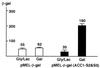
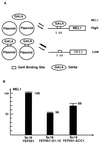
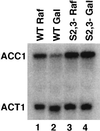
Understanding how a regulatory protein occupies its sites in vivo is central to understanding gene regulation. Using the yeast Gal4 protein as a model for such studies, we have found 239 potential Gal4 binding sites in the yeast genome, 186 of which are in open reading frames (ORFs). This raises the questions of whether these sites are occupied by Gal4 and, if so, to what effect. We have analyzed the Saccharomyces cerevisiae ACC1 gene (encoding acetyl-coenzyme A carboxylase), which has three Gal4 binding sites in its ORF. The plasmid titration assay has demonstrated that Gal4 occupies these sites in the context of an active ACC1 gene. We also find that the expression of the ACC1 is reduced fourfold in galactose medium and that this reduction is dependent on the Gal4 binding sites, suggesting that Gal4 bound to the ORF sites affects transcription of ACC1. Interestingly, removal of the Gal4 binding sites has no obvious effect on the growth in galactose under laboratory conditions. In addition, though the sequence of the ACC1 gene is highly conserved among yeast species, these Gal4 binding sites are not present in the Kluyveromyces lactis ACC1 gene. We suggest that the occurrence of these sites may not be related to galactose regulation and a manifestation of the "noise" in the occurrence of Gal4 binding sites.
Figures




Similar articles
-
How does the GAL4 transcription factor recognize the appropriate DNA binding sites in vivo?Cell Mol Biol Res. 1993;39(4):355-60. Cell Mol Biol Res. 1993. PMID: 8312971
-
Negative effect of the transcriptional activator GAL4.Nature. 1988 Aug 25;334(6184):721-4. doi: 10.1038/334721a0. Nature. 1988. PMID: 3412449
-
Mitochondrial dysfunction enhances Gal4-dependent transcription.FEMS Microbiol Lett. 2005 Dec 15;253(2):207-13. doi: 10.1016/j.femsle.2005.09.033. Epub 2005 Oct 7. FEMS Microbiol Lett. 2005. PMID: 16239078
-
Yeast Gal4: a transcriptional paradigm revisited.EMBO Rep. 2006 May;7(5):496-9. doi: 10.1038/sj.embor.7400679. EMBO Rep. 2006. PMID: 16670683 Free PMC article. Review.
-
[Mechanism of gene-expression regulation by inorganic phosphate in yeast].Tanpakushitsu Kakusan Koso. 1994 Mar;39(4):493-502. Tanpakushitsu Kakusan Koso. 1994. PMID: 8165294 Review. Japanese. No abstract available.
Cited by
-
A downstream regulatory element located within the coding sequence mediates autoregulated expression of the yeast fatty acid synthase gene FAS2 by the FAS1 gene product.Nucleic Acids Res. 2001 Nov 15;29(22):4625-32. doi: 10.1093/nar/29.22.4625. Nucleic Acids Res. 2001. PMID: 11713312 Free PMC article.
-
Transcription-dependent spreading of the Dal80 yeast GATA factor across the body of highly expressed genes.PLoS Genet. 2019 Feb 28;15(2):e1007999. doi: 10.1371/journal.pgen.1007999. eCollection 2019 Feb. PLoS Genet. 2019. PMID: 30818362 Free PMC article.
-
Gcn4 occupancy of open reading frame regions results in the recruitment of chromatin-modifying complexes but not the mediator complex.EMBO Rep. 2003 Sep;4(9):872-6. doi: 10.1038/sj.embor.embor931. EMBO Rep. 2003. PMID: 12949586 Free PMC article.
-
Evolution of biological interaction networks: from models to real data.Genome Biol. 2011 Dec 28;12(12):235. doi: 10.1186/gb-2011-12-12-235. Genome Biol. 2011. PMID: 22204388 Free PMC article. Review.
-
Leucine biosynthesis in fungi: entering metabolism through the back door.Microbiol Mol Biol Rev. 2003 Mar;67(1):1-15, table of contents. doi: 10.1128/MMBR.67.1.1-15.2003. Microbiol Mol Biol Rev. 2003. PMID: 12626680 Free PMC article. Review.
References
-
- Berger S L, Cress W D, Cress A, Triezenberg S J, Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell. 1990;61:1199–208. - PubMed
-
- Breunig K D, Kuger P. Functional homology between the yeast regulatory proteins GAL4 and LAC9: LAC9-mediated transcriptional activation in Kluyveromyces lactis involves protein binding to a regulatory sequence homologous to the GAL4 protein-binding site. Mol Cell Biol. 1987;7:4400–4406. - PMC - PubMed
-
- Chen X, Farmer G, Zhu H, Prywes R, Prives C. Cooperative DNA binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev. 1993;7:1837–1849. - PubMed
-
- Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
