The Sm domain is an ancient RNA-binding motif with oligo(U) specificity
- PMID: 11259661
- PMCID: PMC31112
- DOI: 10.1073/pnas.071033998
The Sm domain is an ancient RNA-binding motif with oligo(U) specificity
Abstract
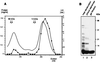
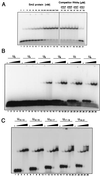
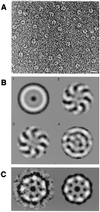
Sm and Sm-like proteins are members of a family of small proteins that is widespread throughout eukaryotic kingdoms. These proteins form heteromers with one another and bind, as heteromeric complexes, to various RNAs, recognizing primarily short U-rich stretches. Interestingly, completion of several genome projects revealed that archaea also contain genes that may encode Sm-like proteins. Herein, we studied the properties of one Sm-like protein derived from the archaebacterium Archaeoglobus fulgidus and overexpressed in Escherichia coli. This single small protein closely reflects the properties of an Sm or Sm-like protein heteromer. It binds to RNA with a high specificity for oligo(U), and assembles onto the RNA to form a complex that exhibits, as judged by electron microscopy, a ring-like structure similar to the ones observed with the Sm core ribonucleoprotein and the like Sm (LSm) protein heteromer. Importantly, multivariate statistical analysis of negative-stain electron-microscopic images revealed a sevenfold symmetry for the observed ring structure, indicating that the proteins form a homoheptamer. These results support the structural model of the Sm proteins derived from crystallographic studies on Sm heterodimers and demonstrate that the Sm protein family evolved from a single ancestor that was present before the eukaryotic and archaeal kingdoms separated.
Figures
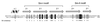
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

