Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome
- PMID: 11259664
- PMCID: PMC31190
- DOI: 10.1073/pnas.071045598
Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome
Abstract
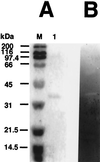
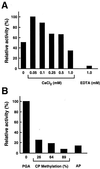
Clostridium cellulovorans uses not only cellulose but also xylan, mannan, pectin, and several other carbon sources for its growth and produces an extracellular multienzyme complex called the cellulosome, which is involved in plant cell wall degradation. Here we report a gene for a cellulosomal subunit, pectate lyase A (PelA), lying downstream of the engY gene, which codes for cellulosomal enzyme EngY. pelA is composed of an ORF of 2,742 bp and encodes a protein of 914 aa with a molecular weight of 94,458. The amino acid sequence derived from pelA revealed a multidomain structure, i.e., an N-terminal domain partially homologous to the C terminus of PelB of Erwinia chrysanthemi belonging to family 1 of pectate lyases, a putative cellulose-binding domain, a catalytic domain homologous to PelL and PelX of E. chrysanthemi that belongs to family 4 of pectate lyases, and a duplicated sequence (or dockerin) at the C terminus that is highly conserved in enzymatic subunits of the C. cellulovorans cellulosome. The recombinant truncated enzyme cleaved polygalacturonic acid to digalacturonic acid (G2) and trigalacturonic acid (G3) but did not act on G2 and G3. There have been no reports available to date on pectate lyase genes from Clostridia.
Figures





References
-
- Albersheim P, Killias U. Arch Biochem Biophys. 1962;97:107–115. - PubMed
-
- Barras F, van Gijsegem F, Chatterjee A K. Annu Rev Phytopathol. 1994;32:201–234.
-
- Collmer A, Keen N T. Annu Rev Phytopathol. 1986;24:383–409.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

