Role of the arginine-nitric oxide pathway in the regulation of vascular smooth muscle cell proliferation
- PMID: 11259671
- PMCID: PMC31203
- DOI: 10.1073/pnas.071054698
Role of the arginine-nitric oxide pathway in the regulation of vascular smooth muscle cell proliferation
Abstract
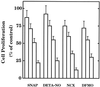
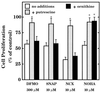
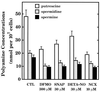
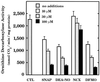
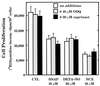
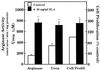
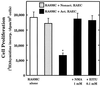
The objective of this study was to elucidate the mechanisms by which nitric oxide (NO) inhibits rat aortic smooth muscle cell (RASMC) proliferation. Two products of the arginine-NO pathway interfere with cell growth by distinct mechanisms. N(G)-hydroxyarginine and NO appear to interfere with cell proliferation by inhibiting arginase and ornithine decarboxylase (ODC), respectively. S-nitroso-N-acetylpenicillamine, (Z)-1-[N-(2-aminoethyl)-N-(2-aminoethyl)-amino]-diazen-1-ium-1,2-diolate, and a nitroaspirin derivative (NCX 4016), each of which is a NO donor agent, inhibited RASMC growth at concentrations of 1-3 microM by cGMP-independent mechanisms. The cytostatic action of the NO donor agents as well as alpha-difluoromethylornithine (DFMO), a known ODC inhibitor, was prevented by addition of putrescine but not ornithine. These observations suggested that NO, like DFMO, may directly inhibit ODC. Experiments with purified, recombinant mammalian ODC revealed that NO inhibits ODC possibly by S-nitrosylation of the active site cysteine in ODC. DFMO, as well as the NO donor agents, interfered with cellular polyamine (putrescine, spermidine, spermine) production. Conversely, increasing the expression and catalytic activity of arginase I in RASMC either by transfection of cells with the arginase I gene or by induction of arginase I mRNA with IL-4 resulted in increased urea and polyamine production as well as cell proliferation. Finally, coculture of rat aortic endothelial cells, which had been pretreated with lipopolysaccharide plus a cytokine mixture to induce NO synthase and promote NO production, caused NO-dependent inhibition of target RASMC proliferation. This study confirms the inhibitory role of the arginine-NO pathway in vascular smooth muscle proliferation and indicates that one mechanism of action of NO is cGMP-independent and attributed to its capacity to inhibit ODC.
Figures


References
-
- Moncada S, Palmer R M J, Higgs E A. Pharmacol Rev. 1991;43:109–142. - PubMed
-
- Buga G M, Wei L H, Bauer P M, Fukuto J M, Ignarro L J. Am J Physiol. 1998;275:R1256–R1264. - PubMed
-
- Blachier F, Robert V, Selamnia M, Mayeur C, Duee P H. FEBS Lett. 1996;396:315–318. - PubMed
-
- Garg U C, Hassid A. Am J Physiol. 1989;257:F60–F66. - PubMed
-
- Garg U C, Hassid A. Biochem Biophys Res Commun. 1990;171:474–479. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

