A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit
- PMID: 11259679
- PMCID: PMC31120
- DOI: 10.1073/pnas.071527498
A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit
Abstract
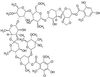
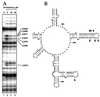
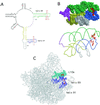
Evernimicin (Evn), an oligosaccharide antibiotic, interacts with the large ribosomal subunit and inhibits bacterial protein synthesis. RNA probing demonstrated that the drug protects a specific set of nucleotides in the loops of hairpins 89 and 91 of 23S rRNA in bacterial and archaeal ribosomes. Spontaneous Evn-resistant mutants of Halobacterium halobium contained mutations in hairpins 89 and 91 of 23S rRNA. In the ribosome tertiary structure, rRNA residues involved in interaction with the drug form a tight cluster that delineates the drug-binding site. Resistance mutations in the bacterial ribosomal protein L16, which is shown to be homologous to archaeal protein L10e, cluster to the same region as the rRNA mutations. The Evn-binding site overlaps with the binding site of initiation factor 2. Evn inhibits activity of initiation factor 2 in vitro, suggesting that the drug interferes with formation of the 70S initiation complex. The site of Evn binding and its mode of action are distinct from other ribosome-targeted antibiotics. This antibiotic target site can potentially be used for the development of new antibacterial drugs.
Figures

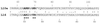
References
-
- Cundliffe E. In: The Molecular Basis of Antibiotic Action. Gale E F, Cundliffe E, Reynolds P E, Richmond M H, Waring M J, editors. New York: Wiley; 1981. pp. 402–545.
-
- Yoshizawa S, Fourmy D, Puglisi J D. Science. 1999;285:1722–1725. - PubMed
-
- Purohit P, Stern S. Nature (London) 1994;370:659–662. - PubMed
-
- Nissen P, Hansen J, Ban N, Moore P B, Steitz T A. Science. 2000;289:920–930. - PubMed
-
- Garrett R A, Rodriguez-Fonseca C. In: Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein Biosynthesis. Zimmermann R A, Dahlberg A E, editors. Boca Raton, FL: CRC; 1996. pp. 327–355.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

