Sequence requirements for Sindbis virus subgenomic mRNA promoter function in cultured cells
- PMID: 11264340
- PMCID: PMC114842
- DOI: 10.1128/JVI.75.8.3509-3519.2001
Sequence requirements for Sindbis virus subgenomic mRNA promoter function in cultured cells
Abstract
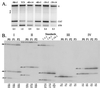
The Sindbis virus minimal subgenomic mRNA promoter (spanning positions -19 to +5 relative to the subgenomic mRNA start site) is approximately three- to sixfold less active than the fully active -98 to +14 promoter region. We identified two elements flanking the -19 to +5 region which increase its transcription to levels comparable to the -98 to +14 region. These elements span positions -40 to -20 and +6 to +14 and act synergistically to enhance transcription. Nine different virus libraries were constructed containing blocks of five randomized nucleotides at various positions in the -40 to +14 region. On passaging these libraries in mosquito cells, a small subset of the viruses came to dominate the population. Sequence analysis at the population level and for individual clones revealed that in general, wild-type bases were preferred for positions -15 to +5 of the minimal promoter. Base mutagenesis experiments indicated that the selection of wild-type bases in this region was primarily due to requirements for subgenomic mRNA transcription. Outside of the minimal promoter, the -35 to -29 region contained four positions which also preferred wildtype bases. However, the remaining positions generally preferred non-wild-type bases. On passaging of the virus libraries on hamster cells, the -15 to +5 region again preferred the wild-type base but most of the remaining positions exhibited almost no base preference. The promoter thus consists of an essential central region from -15 to +5 and discrete flanking sites that render it fully active, depending on the host environment.
Figures


References
-
- Carmichael G G, McMaster G K. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65:380–391. - PubMed
-
- Chang G J, Trent D W. Nucleotide sequence of the genome region encoding the 26S mRNA of eastern equine encephalomyelitis virus and the deduced amino acid sequence of the viral structural proteins. J Gen Virol. 1987;68:2129–2142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

