Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus
- PMID: 11264354
- PMCID: PMC114856
- DOI: 10.1128/JVI.75.8.3647-3656.2001
Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus
Abstract
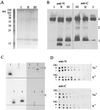
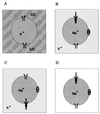
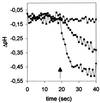
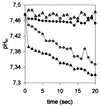
The viral ion channel protein M2 supports the transit of influenza virus and its glycoproteins through acidic compartments of the cell. M2 conducts endosomal protons into the virion to initiate uncoating and, by equilibrating the pH at trans-Golgi membranes, preserves the native conformation of acid-sensitive viral hemagglutinin. The exceptionally low conductance of the M2 channel thwarted resolution of single channels by electrophysiological techniques. Assays of liposome-reconstituted M2 yielded the average unitary channel current of the M2 tetramer--1.2 aA (1.2 x 10(-18) A) at neutral pH and 2.7 to 4.1 aA at pH 5.7--which activates the channel. Extrapolation to physiological temperature predicts 4.8 and 40 aA, respectively, and a unitary conductance of 0.03 versus 0.4 fS. This minute activity, below previous estimates, appears sufficient for virus reproduction, but low enough to avert abortive cytotoxicity. The unitary permeability of M2 was within the range reported for other proton channels. To address the ion selectivity of M2, we exploited the coupling of ionic influx and efflux in sealed liposomes. Metal ion fluxes were monitored by proton counterflow, employing a pH probe 1,000 times more sensitive than available Na+ or K+ probes. Even low-pH-activated M2 did not conduct Na+ and K+. The proton selectivity of M2 was estimated to be at least 3 x 10(6) (over sodium or potassium ions), in agreement with electrophysiological studies. The stringent proton selectivity of M2 suggests that the cytopathology of influenza virus does not involve direct perturbation of cellular sodium or potassium gradients.
Figures

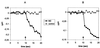
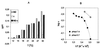
Similar articles
-
Proton transport through influenza A virus M2 protein reconstituted in vesicles.Biophys J. 2008 Jan 15;94(2):434-45. doi: 10.1529/biophysj.107.109082. Epub 2007 Sep 7. Biophys J. 2008. PMID: 17827230 Free PMC article.
-
Ion selectivity and activation of the M2 ion channel of influenza virus.Biophys J. 1996 Mar;70(3):1335-46. doi: 10.1016/S0006-3495(96)79690-X. Biophys J. 1996. PMID: 8785289 Free PMC article.
-
Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells.J Physiol. 1996 Jul 15;494 ( Pt 2)(Pt 2):329-36. doi: 10.1113/jphysiol.1996.sp021495. J Physiol. 1996. PMID: 8841994 Free PMC article.
-
Influenza virus M2 protein and haemagglutinin conformation changes during intracellular transport.Acta Virol. 1995 Jun;39(3):171-81. Acta Virol. 1995. PMID: 8579000 Review.
-
[Structure and function of the influenza virus M2 ion channel protein].Nihon Rinsho. 1997 Oct;55(10):2587-92. Nihon Rinsho. 1997. PMID: 9360376 Review. Japanese.
Cited by
-
Voltage-gated proton channels: molecular biology, physiology, and pathophysiology of the H(V) family.Physiol Rev. 2013 Apr;93(2):599-652. doi: 10.1152/physrev.00011.2012. Physiol Rev. 2013. PMID: 23589829 Free PMC article. Review.
-
Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer.Science. 2010 Oct 22;330(6003):509-12. doi: 10.1126/science.1191750. Science. 2010. PMID: 20966252 Free PMC article.
-
Mechanism of drug inhibition and drug resistance of influenza A M2 channel.Proc Natl Acad Sci U S A. 2009 May 5;106(18):7379-84. doi: 10.1073/pnas.0902548106. Epub 2009 Apr 21. Proc Natl Acad Sci U S A. 2009. PMID: 19383794 Free PMC article.
-
pH-dependent conformation, dynamics, and aromatic interaction of the gating tryptophan residue of the influenza M2 proton channel from solid-state NMR.Biophys J. 2013 Apr 16;104(8):1698-708. doi: 10.1016/j.bpj.2013.02.054. Biophys J. 2013. PMID: 23601317 Free PMC article.
-
Proton transport through influenza A virus M2 protein reconstituted in vesicles.Biophys J. 2008 Jan 15;94(2):434-45. doi: 10.1529/biophysj.107.109082. Epub 2007 Sep 7. Biophys J. 2008. PMID: 17827230 Free PMC article.
References
-
- Aidley D J, Stanfield P R. Ion channels: molecules in action. Cambridge, United Kingdom: Cambridge University Press; 1996.
-
- Beaven G H, Holiday E R. Ultraviolet absorption spectra of proteins and amino acids. Adv Protein Chem. 1952;7:319–368. - PubMed
-
- Ciampor F, Bayley P M, Nermut M V, Hirst E M A, Sugrue R J, Hay A J. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus haemagglutinin to the low pH conformation occurs in an acidic trans-Golgi compartment. Virology. 1992;188:14–24. - PubMed
-
- Ciampor F, Thompson C A, Hay A J. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 1992;22:247–258. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

