Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999
- PMID: 11264359
- PMCID: PMC114861
- DOI: 10.1128/JVI.75.8.3696-3705.2001
Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999
Abstract
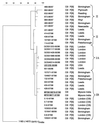
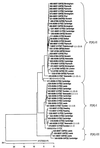
The G and P genotypes of 3,601 rotavirus strains collected in the United Kingdom between 1995 and 1999 were determined (M. Iturriza-Gómara et al., J. Clin. Microbiol. 38:4394-4401, 2000). In 95.4% of the strains the most common G and P combinations, G1P[8], G2P[4], G3P[8], and G4P[8], were found. A small but significant number (2%) of isolates from the remaining strains were reassortants of the most common cocirculating strains, e.g., G1P[4] and G2P[8]. Rotavirus G9P[6] and G9P[8] strains, which constituted 2.7% of all viruses, were genetically closely related in their G components, but the P components of the G9P[8] strains were very closely related to those of cocirculating strains of the more common G types (G1, G3, and G4). In conclusion, genetic interaction by reassortment among cocirculating rotaviruses is not a rare event and contributes significantly to their overall diversity.
Figures


References
-
- Blackhall J, Fuentes A, Magnusson G. Genetic stability of a porcine rotavirus RNA segment during repeated plaque isolation. Virology. 1996;225:181–190. - PubMed
-
- Browning G F, Snodgrass D R, Nakagomi O, Kaga E, Sarasini A, Gerna G. Human and bovine serotype G8 rotaviruses may be derived by reassortment. Arch Virol. 1992;125:121–128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

