CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and T cells
- PMID: 11264367
- PMCID: PMC114869
- DOI: 10.1128/JVI.75.8.3779-3790.2001
CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and T cells
Abstract
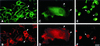
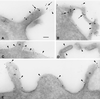
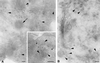
The chemokine receptors CCR5 and CXCR4 act synergistically with CD4 in an ordered multistep mechanism to allow the binding and entry of human immunodeficiency virus type 1 (HIV-1). The efficiency of such a coordinated mechanism depends on the spatial distribution of the participating molecules on the cell surface. Immunoelectron microscopy was performed to address the subcellular localization of the chemokine receptors and CD4 at high resolution. Cells were fixed, cryoprocessed, and frozen; 80-nm cryosections were double labeled with combinations of CCR5, CXCR4, and CD4 antibodies and then stained with immunogold. Surprisingly, CCR5, CXCR4, and CD4 were found predominantly on microvilli and appeared to form homogeneous microclusters in all cell types examined, including macrophages and T cells. Further, while mixed microclusters were not observed, homogeneous microclusters of CD4 and the chemokine receptors were frequently separated by distances less than the diameter of an HIV-1 virion. Such distributions are likely to facilitate cooperative interactions with HIV-1 during virus adsorption to and penetration of human leukocytes and have significant implications for development of therapeutically useful inhibitors of the entry process. Although the mechanism underlying clustering is not understood, clusters were observed in small trans-Golgi vesicles, implying that they were organized shortly after synthesis and well before insertion into the cellular membrane. Chemokine receptors normally act as sensors, detecting concentration gradients of their ligands and thus providing directional information for cellular migration during both normal homeostasis and inflammatory responses. Localization of these sensors on the microvilli should enable more precise monitoring of their environment, improving efficiency of the chemotactic process. Moreover, since selectins, some integrins, and actin are also located on or in the microvillus, this organelle has many of the major elements required for chemotaxis.
Figures






References
-
- Aithal H N, Knigge K M, Kartha S, Czyzewski E A, Toback F G. An alternate method utilizing small quantities of ligand for affinity purification of monospecific antibodies. J Immunol Methods. 1988;112:63–69. - PubMed
-
- Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. - PubMed
-
- Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol. 1997;15:675–705. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

