Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1
- PMID: 11264372
- PMCID: PMC114874
- DOI: 10.1128/JVI.75.8.3832-3840.2001
Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1
Abstract
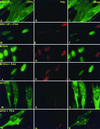
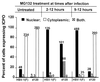
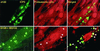
Earlier studies have shown that wild-type infected-cell protein 0 (ICP0), a key herpes simplex virus regulatory protein, translocates from the nucleus to the cytoplasm of human embryonic lung (HEL) fibroblasts within several hours after infection (Y. Kawaguchi, R. Bruni, and B. Roizman, J. Virol. 71:1019-1024, 1997). Translocation of ICP0 was also observed in cells infected with the d120 mutant, in which both copies of the gene encoding ICP4, the major regulatory protein, had been deleted (V. Galvan, R. Brandimarti, J. Munger, and B. Roizman, J. Virol. 74:1931-1938, 2000). Furthermore, a mutant (R7914) carrying the D199A substitution in ICP0 does not bind or stabilize cyclin D3 and is retained in the nucleus (C. Van Sant, P. Lopez, S. J. Advani, and B. Roizman, J. Virol. 75:1888-1898, 2001). Studies designed to elucidate the requirements for the translocation of ICP0 between cellular compartments revealed the following. (i) Translocation of ICP0 to the cytoplasm in productive infection maps to the D199 amino acid, inasmuch as wild-type ICP0 delivered in trans to cells infected with an ICP0 null mutant was translocated to the cytoplasm whereas the D199A-substituted mutant ICP0 was not. (ii) Translocation of wild-type ICP0 requires a function expressed late in infection, inasmuch as phosphonoacetate blocked the translocation of ICP0 in wild-type virus-infected cells but not in d120 mutant-infected cells. Moreover, whereas in d120 mutant-infected cells ICP0 was translocated rapidly from the cytoplasm to the nucleus at approximately 5 h after infection, the translocation of ICP0 in wild-type virus-infected cells extended from 5 to at least 9 h after infection. (iii) In wild-type virus-infected cells, the MG132 proteasomal inhibitor blocked the translocation of ICP0 to the cytoplasm early in infection, but when added late in infection, it caused ICP0 to be relocated back to the nucleus from the cytoplasm. (iv) MG132 blocked the translocation of ICP0 in d120 mutant-infected cells early in infection but had no effect on the ICP0 aggregated in vesicle-like structures late in infection. However, in d120 mutant-infected cells treated with MG132 at late times, proteasomes formed a shell-like structure around the aggregated ICP0. These structures were not seen in wild-type virus or R7914 mutant-infected cells. The results indicate the following. (i) In the absence of beta or gamma protein synthesis, ICP0 dynamically associates with proteasomes and is translocated to the cytoplasm. (ii) In cells productively infected beyond alpha gene expression, ICP0 is retained in the nucleus until after the onset of viral DNA synthesis and the synthesis of gamma2 proteins. (iii) Late in infection, ICP0 is actively sequestered in the cytoplasm by a process mediated by proteasomes, inasmuch as interference with proteasomal function causes rapid relocation of ICP0 to the nucleus.
Figures
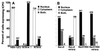


References
-
- Blaho J A, Mitchell C, Roizman B. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J Biol Chem. 1994;269:17401–17410. - PubMed
-
- Chelbi-Alix K M, de The H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935–941. - PubMed
-
- Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

