Fate of mat1 DNA strands during mating-type switching in fission yeast
- PMID: 11265754
- PMCID: PMC1084255
- DOI: 10.1093/embo-reports/kvd023
Fate of mat1 DNA strands during mating-type switching in fission yeast
Abstract
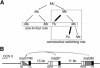
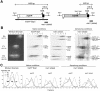
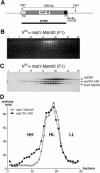
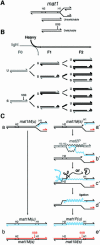
The mating-type switching of the fission yeast, Schizosaccharomyces pombe, is highly regulated. Two consecutive asymmetric divisions are required to produce one mating-type switched cell among the four progeny. Using DNA density-gradient centrifugation we demonstrate that one-fourth of the mat1 DNA is not replicated by the conventional semi-conservative mode, but instead both DNA strands are synthesized de novo. Our data are consistent with a gene conversion event, initiated by a site- and strand-specific DNA break (SSB). We further demonstrate that the virgin switched mat1-containing chromatid no longer contained the nick, while it is reintroduced during the lagging strand synthesis of the mat1 locus on the sister chromatid. This finding establishes at the molecular level a firm experimental link between the phenotype and genotype in the process of asymmetric mating-type switching during mitotic divisions.
Figures
References
-
- Barbet N.C. and Carr, A.M. (1993) Fission yeast wee1 protein kinase is not required for 5 DNA damage-dependent mitotic arrest. Nature, 364, 824–827. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

