The 3.7 A projection map of the glycerol facilitator GlpF: a variant of the aquaporin tetramer
- PMID: 11265760
- PMCID: PMC1084254
- DOI: 10.1093/embo-reports/kvd022
The 3.7 A projection map of the glycerol facilitator GlpF: a variant of the aquaporin tetramer
Abstract
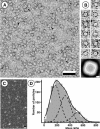
GlpF, the glycerol facilitator protein of Escherichia coli, is an archetypal member of the aquaporin superfamily. To assess its structure, recombinant histidine-tagged protein was overexpressed, solubilized in octylglucoside and purified to homogeneity. Negative stain electron microscopy of solubilized GlpF protein revealed a tetrameric structure of approximately 80 A side length. Scanning transmission electron microscopy yielded a mass of 170 kDa, corroborating the tetrameric nature of GlpF. Reconstitution of GlpF in the presence of lipids produced highly ordered two-dimensional crystals, which diffracted electrons to 3.6 A resolution. Cryoelectron microscopy provided a 3.7 A projection map exhibiting a unit cell comprised of two tetramers. In projection, GlpF is similar to AQP1, the erythrocyte water channel. However, the major density minimum within each monomer is distinctly larger in GlpF than in AQP1.
Figures



Similar articles
-
Reconstitution and functional comparison of purified GlpF and AqpZ, the glycerol and water channels from Escherichia coli.Proc Natl Acad Sci U S A. 2001 Feb 27;98(5):2888-93. doi: 10.1073/pnas.051628098. Epub 2001 Feb 20. Proc Natl Acad Sci U S A. 2001. PMID: 11226336 Free PMC article.
-
The 6.9-A structure of GlpF: a basis for homology modeling of the glycerol channel from Escherichia coli.J Struct Biol. 2000 Nov;132(2):133-41. doi: 10.1006/jsbi.2000.4317. J Struct Biol. 2000. PMID: 11162735
-
Structure of the water channel AqpZ from Escherichia coli revealed by electron crystallography.J Mol Biol. 1999 Sep 3;291(5):1181-90. doi: 10.1006/jmbi.1999.3031. J Mol Biol. 1999. PMID: 10518953
-
The structure of GlpF, a glycerol conducting channel.Novartis Found Symp. 2002;245:51-61; discussion 61-5, 165-8. Novartis Found Symp. 2002. PMID: 12027015 Review.
-
Water and glycerol permeation through the glycerol channel GlpF and the aquaporin family.J Synchrotron Radiat. 2004 Jan 1;11(Pt 1):86-8. doi: 10.1107/s0909049503023872. Epub 2003 Nov 28. J Synchrotron Radiat. 2004. PMID: 14646142 Review.
Cited by
-
Aquaporin Gating: A New Twist to Unravel Permeation through Water Channels.Int J Mol Sci. 2022 Oct 14;23(20):12317. doi: 10.3390/ijms232012317. Int J Mol Sci. 2022. PMID: 36293170 Free PMC article. Review.
-
Molecular dissection of water and glycerol permeability of the aquaglyceroporin from Plasmodium falciparum by mutational analysis.Proc Natl Acad Sci U S A. 2004 Feb 3;101(5):1153-8. doi: 10.1073/pnas.0307295101. Epub 2004 Jan 20. Proc Natl Acad Sci U S A. 2004. PMID: 14734807 Free PMC article.
-
Identification of key residues involved in Si transport by the aquaglyceroporins.J Gen Physiol. 2016 Sep;148(3):239-51. doi: 10.1085/jgp.201611598. Epub 2016 Aug 15. J Gen Physiol. 2016. PMID: 27527099 Free PMC article.
-
Reconstitution and functional comparison of purified GlpF and AqpZ, the glycerol and water channels from Escherichia coli.Proc Natl Acad Sci U S A. 2001 Feb 27;98(5):2888-93. doi: 10.1073/pnas.051628098. Epub 2001 Feb 20. Proc Natl Acad Sci U S A. 2001. PMID: 11226336 Free PMC article.
-
Projection map of aquaporin-9 at 7 A resolution.J Mol Biol. 2007 Mar 16;367(1):80-8. doi: 10.1016/j.jmb.2006.12.042. Epub 2006 Dec 20. J Mol Biol. 2007. PMID: 17239399 Free PMC article.
References
-
- Baumeister W., Karrenberg, F., Rachel, R., Engel, A., ten Heggeler, B. and Saxton, W.O. (1982) The major cell envelope protein of Micrococcus radiodurans (R1). Structural and chemical characterization. Eur. J. Biochem., 125, 535–544. - PubMed
-
- Bevington P.R. (1969) Data Reduction and Error Analysis for the Physical Sciences. McGraw–Hill, New York, NY.
-
- Borgnia M.J., Kozono, D., Calamita, G., Maloney, P.C. and Agre, P. (1999) Functional reconstitution and characterization of AqpZ, the E. coli water channel protein. J. Mol. Biol., 291, 1169–1179. - PubMed
-
- Bron P. et al. (1999) Oligomerization state of MIP proteins expressed in Xenopus oocytes as revealed by freeze-fracture electron-microscopy analysis. J. Struct. Biol., 128, 287–296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

