The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation
- PMID: 11266451
- PMCID: PMC2199619
- DOI: 10.1083/jcb.152.2.349
The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation
Abstract
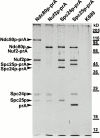
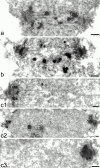
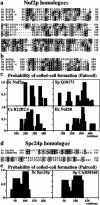
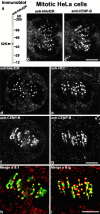
We have purified a complex from Saccharomyces cerevisiae containing the spindle components Ndc80p, Nuf2p, Spc25p, and Spc24p. Temperature-sensitive mutants in NDC80, SPC25, and SPC24 show defects in chromosome segregation. In spc24-1 cells, green fluorescence protein (GFP)-labeled centromeres fail to split during spindle elongation, and in addition some centromeres may detach from the spindle. Chromatin immunoprecipitation assays show an association of all four components of the complex with the yeast centromere. Homologues of Ndc80p, Nuf2p, and Spc24p were found in Schizosaccharomyces pombe and GFP tagging showed they were located at the centromere. A human homologue of Nuf2p was identified in the expressed sequence tag database. Immunofluorescent staining with anti-human Nuf2p and with anti-HEC, the human homologue of Ndc80p, showed that both proteins are at the centromeres of mitotic HeLa cells. Thus the Ndc80p complex contains centromere-associated components conserved between yeasts and vertebrates.
Figures





References
-
- Adams R.R., Wheatley S.P., Gouldsworthy A.M., Kandels-Lewis S.E., Carmena M., Smythe C., Gerloff D.L., Earnshaw W.C. INCENP binds the aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000;10:1075–1078. - PubMed
-
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. Two novel related yeast nucleoporins Nup170p and Nup155pcomplementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1995;131:1133–1148. - PMC - PubMed
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Bahler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe . Yeast. 1998;14:943–951. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

