Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability
- PMID: 11266455
- PMCID: PMC2199622
- DOI: 10.1083/jcb.152.2.401
Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability
Abstract
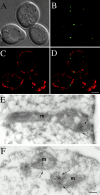
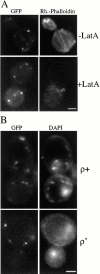
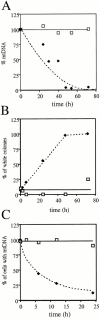
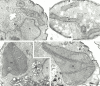
In the yeast Saccharomyces cerevisiae, mitochondria form a branched, tubular reticulum in the periphery of the cell. Mmm1p is required to maintain normal mitochondrial shape and in mmm1 mutants mitochondria form large, spherical organelles. To further explore Mmm1p function, we examined the localization of a Mmm1p-green fluorescent protein (GFP) fusion in living cells. We found that Mmm1p-GFP is located in small, punctate structures on the mitochondrial outer membrane, adjacent to a subset of matrix-localized mitochondrial DNA nucleoids. We also found that the temperature-sensitive mmm1-1 mutant was defective in transmission of mitochondrial DNA to daughter cells immediately after the shift to restrictive temperature. Normal mitochondrial nucleoid structure also collapsed at the nonpermissive temperature with similar kinetics. Moreover, we found that mitochondrial inner membrane structure is dramatically disorganized in mmm1 disruption strains. We propose that Mmm1p is part of a connection between the mitochondrial outer and inner membranes, anchoring mitochondrial DNA nucleoids in the matrix.
Figures




References
-
- Adams A., Gottschling D.E., Kaiser C.A., Stearns T. Methods in Yeast Genetics 1997. Cold Spring Harbor Laboratories; Cold Spring Harbor, N.Y: pp. 1–112
-
- Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int. Rev. Cytol. 1990;122:1–62. - PubMed
-
- Bereiter-Hahn J., Voth M. Dynamics of mitochondria in living cellsshape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 1994;27:198–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

